Ce-emerin and LEM-2: essential roles in Caenorhabditis elegans development, muscle function, and mitosis
- PMID: 22171324
- PMCID: PMC3279384
- DOI: 10.1091/mbc.E11-06-0505
Ce-emerin and LEM-2: essential roles in Caenorhabditis elegans development, muscle function, and mitosis
Abstract
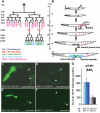
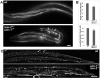
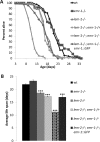
Emerin and LEM2 are ubiquitous inner nuclear membrane proteins conserved from humans to Caenorhabditis elegans. Loss of human emerin causes Emery-Dreifuss muscular dystrophy (EDMD). To test the roles of emerin and LEM2 in somatic cells, we used null alleles of both genes to generate C. elegans animals that were either hypomorphic (LEM-2-null and heterozygous for Ce-emerin) or null for both proteins. Single-null and hypomorphic animals were viable and fertile. Double-null animals used the maternal pool of Ce-emerin to develop to the larval L2 stage, then arrested. Nondividing somatic cell nuclei appeared normal, whereas dividing cells had abnormal nuclear envelope and chromatin organization and severe defects in postembryonic cell divisions, including the mesodermal lineage. Life span was unaffected by loss of Ce-emerin alone but was significantly reduced in LEM-2-null animals, and double-null animals had an even shorter life span. In addition to striated muscle defects, double-null animals and LEM-2-null animals showed unexpected defects in smooth muscle activity. These findings implicate human LEM2 mutations as a potential cause of EDMD and further suggest human LEM2 mutations might cause distinct disorders of greater severity, since C. elegans lacking only LEM-2 had significantly reduced life span and smooth muscle activity.
Figures





References
-
- Bione S, Maestrini E, Rivella S, Mancini M, Regis S, Romeo G, Toniolo D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet. 1994;8:323–327. - PubMed
-
- Cohen M, Tzur YB, Neufeld E, Feinstein N, Delannoy MR, Wilson KL, Gruenbaum Y. Transmission electron microscope studies of the nuclear envelope in Caenorhabditis elegans embryos. J Struct Biol. 2002;140:232–240. - PubMed
-
- Corsi AK, Kostas SA, Fire A, Krause M. Caenorhabditis elegans Twist plays an essential role in non-striated muscle development. Development. 2000;127:2041–2051. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

