The mosquito Aedes aegypti has a large genome size and high transposable element load but contains a low proportion of transposon-specific piRNAs
- PMID: 22171608
- PMCID: PMC3259105
- DOI: 10.1186/1471-2164-12-606
The mosquito Aedes aegypti has a large genome size and high transposable element load but contains a low proportion of transposon-specific piRNAs
Abstract
Background: The piRNA pathway has been shown in model organisms to be involved in silencing of transposons thereby providing genome stability. In D. melanogaster the majority of piRNAs map to these sequences. The medically important mosquito species Aedes aegypti has a large genome size, a high transposon load which includes Miniature Inverted repeat Transposable Elements (MITES) and an expansion of the piRNA biogenesis genes. Studies of transgenic lines of Ae. aegypti have indicated that introduced transposons are poorly remobilized and we sought to explore the basis of this. We wished to analyze the piRNA profile of Ae. aegypti and thereby determine if it is responsible for transposon silencing in this mosquito.
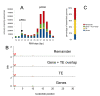
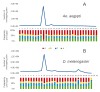
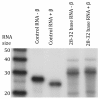
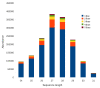
Results: Estimated piRNA sequence diversity was comparable between Ae. aegypti and D. melanogaster, but surprisingly only 19% of mosquito piRNAs mapped to transposons compared to 51% for D. melanogaster. Ae. aegypti piRNA clusters made up a larger percentage of the total genome than those of D. melanogaster but did not contain significantly higher percentages of transposon derived sequences than other regions of the genome. Ae. aegypti contains a number of protein coding genes that may be sources of piRNA biogenesis with two, traffic jam and maelstrom, implicated in this process in model organisms. Several genes of viral origin were also targeted by piRNAs. Examination of six mosquito libraries that had previously been transformed with transposon derived sequence revealed that new piRNA sequences had been generated to the transformed sequences, suggesting that they may have stimulated a transposon inactivation mechanism.
Conclusions: Ae. aegypti has a large piRNA complement that maps to transposons but primarily gene sequences, including many viral-derived sequences. This, together the more uniform distribution of piRNA clusters throughout its genome, suggest that some aspects of the piRNA system differ between Ae. aegypti and D. melanogaster.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

