Performance analysis of novel methods for detecting epistasis
- PMID: 22172045
- PMCID: PMC3259123
- DOI: 10.1186/1471-2105-12-475
Performance analysis of novel methods for detecting epistasis
Abstract
Background: Epistasis is recognized fundamentally important for understanding the mechanism of disease-causing genetic variation. Though many novel methods for detecting epistasis have been proposed, few studies focus on their comparison. Undertaking a comprehensive comparison study is an urgent task and a pathway of the methods to real applications.
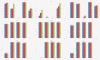
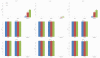
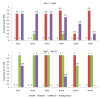
Results: This paper aims at a comparison study of epistasis detection methods through applying related software packages on datasets. For this purpose, we categorize methods according to their search strategies, and select five representative methods (TEAM, BOOST, SNPRuler, AntEpiSeeker and epiMODE) originating from different underlying techniques for comparison. The methods are tested on simulated datasets with different size, various epistasis models, and with/without noise. The types of noise include missing data, genotyping error and phenocopy. Performance is evaluated by detection power (three forms are introduced), robustness, sensitivity and computational complexity.
Conclusions: None of selected methods is perfect in all scenarios and each has its own merits and limitations. In terms of detection power, AntEpiSeeker performs best on detecting epistasis displaying marginal effects (eME) and BOOST performs best on identifying epistasis displaying no marginal effects (eNME). In terms of robustness, AntEpiSeeker is robust to all types of noise on eME models, BOOST is robust to genotyping error and phenocopy on eNME models, and SNPRuler is robust to phenocopy on eME models and missing data on eNME models. In terms of sensitivity, AntEpiSeeker is the winner on eME models and both SNPRuler and BOOST perform well on eNME models. In terms of computational complexity, BOOST is the fastest among the methods. In terms of overall performance, AntEpiSeeker and BOOST are recommended as the efficient and effective methods. This comparison study may provide guidelines for applying the methods and further clues for epistasis detection.
Figures

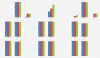
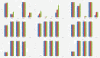
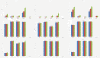

Similar articles
-
An empirical comparison of several recent epistatic interaction detection methods.Bioinformatics. 2011 Nov 1;27(21):2936-43. doi: 10.1093/bioinformatics/btr512. Epub 2011 Sep 7. Bioinformatics. 2011. PMID: 21903628
-
Evaluating the detection ability of a range of epistasis detection methods on simulated data for pure and impure epistatic models.PLoS One. 2022 Feb 18;17(2):e0263390. doi: 10.1371/journal.pone.0263390. eCollection 2022. PLoS One. 2022. PMID: 35180244 Free PMC article.
-
Introducing Heuristic Information Into Ant Colony Optimization Algorithm for Identifying Epistasis.IEEE/ACM Trans Comput Biol Bioinform. 2020 Jul-Aug;17(4):1253-1261. doi: 10.1109/TCBB.2018.2879673. Epub 2018 Nov 5. IEEE/ACM Trans Comput Biol Bioinform. 2020. PMID: 30403637
-
Travelling the world of gene-gene interactions.Brief Bioinform. 2012 Jan;13(1):1-19. doi: 10.1093/bib/bbr012. Epub 2011 Mar 26. Brief Bioinform. 2012. PMID: 21441561 Review.
-
Robust genetic interaction analysis.Brief Bioinform. 2019 Mar 25;20(2):624-637. doi: 10.1093/bib/bby033. Brief Bioinform. 2019. PMID: 29897421 Free PMC article. Review.
Cited by
-
KNN-MDR: a learning approach for improving interactions mapping performances in genome wide association studies.BMC Bioinformatics. 2017 Mar 21;18(1):184. doi: 10.1186/s12859-017-1599-7. BMC Bioinformatics. 2017. PMID: 28327091 Free PMC article.
-
CINOEDV: a co-information based method for detecting and visualizing n-order epistatic interactions.BMC Bioinformatics. 2016 May 17;17(1):214. doi: 10.1186/s12859-016-1076-8. BMC Bioinformatics. 2016. PMID: 27184783 Free PMC article.
-
Confronting the missing epistasis problem: on the reproducibility of gene-gene interactions.Hum Genet. 2015 Aug;134(8):837-49. doi: 10.1007/s00439-015-1564-3. Epub 2015 May 22. Hum Genet. 2015. PMID: 25998948
-
GWIS--model-free, fast and exhaustive search for epistatic interactions in case-control GWAS.BMC Genomics. 2013;14 Suppl 3(Suppl 3):S10. doi: 10.1186/1471-2164-14-S3-S10. Epub 2013 May 28. BMC Genomics. 2013. PMID: 23819779 Free PMC article.
-
Toxo: a library for calculating penetrance tables of high-order epistasis models.BMC Bioinformatics. 2020 Apr 9;21(1):138. doi: 10.1186/s12859-020-3456-3. BMC Bioinformatics. 2020. PMID: 32272874 Free PMC article.
References
-
- Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2(2):91–99. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

