HPV E7 viral oncoprotein disrupts transcriptional regulation of L1Md retrotransposon
- PMID: 22172279
- PMCID: PMC3263525
- DOI: 10.1016/j.febslet.2011.12.005
HPV E7 viral oncoprotein disrupts transcriptional regulation of L1Md retrotransposon
Abstract
Murine L1Md-A5 retrotransposon is a redox-inducible element regulated by Nrf-2/JunD and E2F/Rb-binding sites within its promoter (5'-UTR). Because the human papillomavirus (HPV) oncoprotein E7 interacts with retinoblastoma (pRb) and members of the AP1 family, studies were conducted to examine functional interactions between HPV E7, pRb, and histone deacetylase 2 (HDAC2) in the regulation of L1Md-A5. Using a transient heterologous transcription system we found that HPV E7 alone, or in combination with HDAC2, disrupted pRb-mediated L1MdA-5 transactivation. HPV E7 also ablated the transcriptional response of L1Md-A5 to genotoxic stress, but did not interfere with basal activity. We conclude that HPV E7 associates with proteins involved in the assembly of macromolecular complexes that regulate antioxidant and E2F/Rb sites within L1MdA-5 to regulate biological activity.
Copyright © 2011. Published by Elsevier B.V.
Conflict of interest statement
COMPETING INTEREST: The authors declare no potential competing interests.
Figures

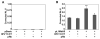
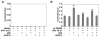

Similar articles
-
The relative ability of human papillomavirus type 6 and human papillomavirus type 16 E7 proteins to transactivate E2F-responsive elements is promoter- and cell-dependent.Virology. 1997 Dec 8;239(1):238-46. doi: 10.1006/viro.1997.8885. Virology. 1997. PMID: 9426463
-
Human papillomavirus type 16 E7 oncoprotein associates with E2F6.J Virol. 2008 Sep;82(17):8695-705. doi: 10.1128/JVI.00579-08. Epub 2008 Jun 25. J Virol. 2008. PMID: 18579589 Free PMC article.
-
Papillomavirus E7 protein binding to the retinoblastoma protein is not required for viral induction of warts.J Virol. 1993 Feb;67(2):716-25. doi: 10.1128/JVI.67.2.716-725.1993. J Virol. 1993. PMID: 8380462 Free PMC article.
-
[Possible role of transcription factor AP1 in the tissue-specific regulation of human papillomavirus].Rev Invest Clin. 2002 May-Jun;54(3):231-42. Rev Invest Clin. 2002. PMID: 12183893 Review. Spanish.
-
Interactions of HPV E6 and E7 oncoproteins with tumour suppressor gene products.Cancer Surv. 1992;12:197-217. Cancer Surv. 1992. PMID: 1322242 Review.
Cited by
-
LINE-1 silencing by retinoblastoma proteins is effected through the nucleosomal and remodeling deacetylase multiprotein complex.BMC Cancer. 2016 Jan 25;16:38. doi: 10.1186/s12885-016-2068-9. BMC Cancer. 2016. PMID: 26810492 Free PMC article.
References
-
- Hardies SC, Martin SL, Voliva CF, Hutchison CA, Edgell MH. An analysis of replacement and synonymous changes in the rodent L1 repeat family. Molecular Biology and Evolution. 1986;3:109–125. - PubMed
-
- Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. MCB.00332-07 [pii] - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

