Digital gene expression profiling of primary acute lymphoblastic leukemia cells
- PMID: 22173241
- PMCID: PMC3377998
- DOI: 10.1038/leu.2011.358
Digital gene expression profiling of primary acute lymphoblastic leukemia cells
Abstract
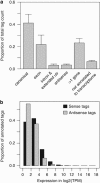
We determined the genome-wide digital gene expression (DGE) profiles of primary acute lymphoblastic leukemia (ALL) cells from 21 patients taking advantage of 'second-generation' sequencing technology. Patients included in this study represent four cytogenetically distinct subtypes of B-cell precursor (BCP) ALL and T-cell lineage ALL (T-ALL). The robustness of DGE combined with supervised classification by nearest shrunken centroids (NSC) was validated experimentally and by comparison with published expression data for large sets of ALL samples. Genes that were differentially expressed between BCP ALL subtypes were enriched to distinct signaling pathways with dic(9;20) enriched to TP53 signaling, t(9;22) to interferon signaling, as well as high hyperdiploidy and t(12;21) to apoptosis signaling. We also observed antisense tags expressed from the non-coding strand of ~50% of annotated genes, many of which were expressed in a subtype-specific pattern. Antisense tags from 17 gene regions unambiguously discriminated between the BCP ALL and T-ALL subtypes, and antisense tags from 76 gene regions discriminated between the 4 BCP subtypes. We observed a significant overlap of gene regions with alternative polyadenylation and antisense transcription (P<1 × 10(-15)). Our study using DGE profiling provided new insights into the RNA expression patterns in ALL cells.
Figures




References
-
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. - PubMed
-
- Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. - PubMed
-
- Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–2959. - PubMed
-
- Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJ, Kazemier KM, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351:533–542. - PubMed
-
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

