Use of a Drosophila genome-wide conserved sequence database to identify functionally related cis-regulatory enhancers
- PMID: 22174086
- PMCID: PMC3243966
- DOI: 10.1002/dvdy.22728
Use of a Drosophila genome-wide conserved sequence database to identify functionally related cis-regulatory enhancers
Abstract
Background: Phylogenetic footprinting has revealed that cis-regulatory enhancers consist of conserved DNA sequence clusters (CSCs). Currently, there is no systematic approach for enhancer discovery and analysis that takes full-advantage of the sequence information within enhancer CSCs.
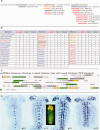
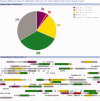
Results: We have generated a Drosophila genome-wide database of conserved DNA consisting of >100,000 CSCs derived from EvoPrints spanning over 90% of the genome. cis-Decoder database search and alignment algorithms enable the discovery of functionally related enhancers. The program first identifies conserved repeat elements within an input enhancer and then searches the database for CSCs that score highly against the input CSC. Scoring is based on shared repeats as well as uniquely shared matches, and includes measures of the balance of shared elements, a diagnostic that has proven to be useful in predicting cis-regulatory function. To demonstrate the utility of these tools, a temporally-restricted CNS neuroblast enhancer was used to identify other functionally related enhancers and analyze their structural organization.
Conclusions: cis-Decoder reveals that co-regulating enhancers consist of combinations of overlapping shared sequence elements, providing insights into the mode of integration of multiple regulating transcription factors. The database and accompanying algorithms should prove useful in the discovery and analysis of enhancers involved in any developmental process.
Copyright © 2011 Wiley Periodicals, Inc.
Figures







References
-
- Alonso ME, Pernaute B, Crespo M, Gómez-Skarmeta JL, Manzanares M. Understanding the regulatory genome. Int J Dev Biol. 2009;53:1367–1378. - PubMed
-
- Anderson MG, Perkins GL, Chittick P, Shrigley RJ, Johnson WA. drifter, a Drosophila POU-domain transcription factor, is required for correct differentiation and migration of tracheal cells and midline glia. Genes Dev. 1995;9:123–137. - PubMed
-
- Armstrong JD, Kaiser K. The study of Drosophila brain development. In: Houdebine LM, editor. Transgenic animals: generation and use. Reading, UK: Harwood Academic Publishers; 1997. pp. 365–370.
-
- Bailey AM, Posakony JW. Suppressor of Hairless directly activates transcription of Enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

