Intrinsic disorder within and flanking the DNA-binding domains of human transcription factors
- PMID: 22174267
- PMCID: PMC3327284
Intrinsic disorder within and flanking the DNA-binding domains of human transcription factors
Abstract
While the term 'protein structure' is commonplace, it is increasingly appreciated that proteins may not possess a single, well-defined structure: some regions of proteins are intrinsically disordered. The role these intrinsically disordered regions (IDRs) play in protein function is an area of significant interest. In particular, because proteins containing IDRs are largely involved in processes related to molecular recognition, the question arises whether IDRs are important in these recognition events. It has been observed that IDRs are enriched in transcription factors (TFs) in comparison with other proteins, and we sought to explore this enrichment more precisely, with an eye toward functional dissection of the prevalence and locations of IDRs in different classes of TFs. Specifically, we considered the occurrences of 76 classes of DNA-binding domains (DBDs) within a comprehensive set of 1,747 human, sequence-specific TFs. For each DBD class, we analyzed whether a significant level of disorder was present within the DBD itself, the N-terminal or C-terminal sequence flanking the DBD, or both flanking sequences. We found that although the DBDs themselves exhibit significant order, the regions flanking the DBDs exhibit significant disorder, which suggests a functional role for such IDRs in TF DNA binding. These results may have important implications for studies of TFs not just in human but across all eukaryotes, and suggest future studies focused on testing the roles of N- and C-terminal flanking regions in determining or modulating the DNA binding affinity and/or specificity of the associated TFs.
Figures
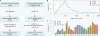

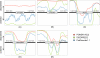
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

