DNA sequence analysis of South African Helicobacter pylori Vacuolating Cytotoxin Gene (vacA)
- PMID: 22174610
- PMCID: PMC3233416
- DOI: 10.3390/ijms12117459
DNA sequence analysis of South African Helicobacter pylori Vacuolating Cytotoxin Gene (vacA)
Abstract
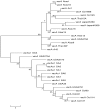
Sequence diversity and population structures can vary widely among pathogenic bacteria species. In some species, all isolates are highly similar, whereas in others most of the isolates are distinguished easily. H. pylori is known for its wide genetic diversity amongst the various strains most especially in the genes involved in virulence. The aim of this study was to evaluate by PCR and sequence analysis, the genetic profile of H. pylori vacA gene (s1, s2, m1 and m2). We sequenced small DNA segments from 13 vacAs1, 10 vacAm2, 6 vacAm1 and 6 vacAs2 strains which were amplified with amplicon size of 259/286 bp, 290 bp and 352 bp for vacAs1/s2, m1 and m2 respectively. Based on similarities among our strains accession numbers were provided for seven vacAs1 (HQ709109-HQ709115), six vacAs2 (JN848463-JN848468), six vacAm1 (JN848469-JN848474) and six vacAm2 (HQ650801-HQ650806) strains. Amongst the strains studied, 98.07%, 98.58%, 97.38% and 95.41% of vacAs1, vacAs2, vacAm1 and vacAm2 of the strains were conserved respectively. Findings of this study underscores the importance of understanding the virulence composition and diversity of H. pylori in South Africa for enhanced clinico-epidemiological monitoring and pathophysiology of disease.
Keywords: Helicobacter pylori; South Africa; diversity; vacuolating cytotoxin gene (vacA).
Figures

Similar articles
-
[The heterogeneity of genetic variants of Helicobacter pylori in patients with various acid-related diseases].Eksp Klin Gastroenterol. 2014;(11):8-13. Eksp Klin Gastroenterol. 2014. PMID: 25842656 Russian.
-
Relationship between Helicobacter pylori virulence genes and clinical outcomes in Saudi patients.J Korean Med Sci. 2012 Feb;27(2):190-3. doi: 10.3346/jkms.2012.27.2.190. Epub 2012 Jan 27. J Korean Med Sci. 2012. PMID: 22323867 Free PMC article.
-
Prevalence of vacA, cagA and babA2 genes in Cuban Helicobacter pylori isolates.World J Gastroenterol. 2009 Jan 14;15(2):204-10. doi: 10.3748/wjg.15.204. World J Gastroenterol. 2009. PMID: 19132771 Free PMC article.
-
Vacuolating cytotoxin A (VacA) - A multi-talented pore-forming toxin from Helicobacter pylori.Toxicon. 2016 Aug;118:27-35. doi: 10.1016/j.toxicon.2016.04.037. Epub 2016 Apr 20. Toxicon. 2016. PMID: 27105670 Review.
-
Influence of Helicobacter pylori virulence factors CagA and VacA on pathogenesis of gastrointestinal disorders.Microb Pathog. 2018 Apr;117:43-48. doi: 10.1016/j.micpath.2018.02.016. Epub 2018 Feb 9. Microb Pathog. 2018. PMID: 29432909 Review.
Cited by
-
Diversification of the vacAs1m1 and vacAs2m2 Strains of Helicobacter pylori in Meriones unguiculatus.Front Microbiol. 2016 Nov 8;7:1758. doi: 10.3389/fmicb.2016.01758. eCollection 2016. Front Microbiol. 2016. PMID: 27877163 Free PMC article.
-
No Association between CagA- and VacA-Positive Strains of Helicobacter pylori and Primary Open-Angle Glaucoma: A Case-Control Study.Ophthalmol Eye Dis. 2016 Feb 17;8:1-4. doi: 10.4137/OED.S35895. eCollection 2016. Ophthalmol Eye Dis. 2016. PMID: 26917977 Free PMC article.
-
Antimicrobial effect of 7-O-butylnaringenin, a novel flavonoid, and various natural flavonoids against Helicobacter pylori strains.Int J Environ Res Public Health. 2013 Oct 28;10(11):5459-69. doi: 10.3390/ijerph10115459. Int J Environ Res Public Health. 2013. PMID: 24169409 Free PMC article.
References
-
- Smith S.I., Kirsch C., Oyedeji K.S., Arigbabu A.O., Coker A.O., Bayerdöffer E., Miehlke S. Prevalence of Helicobacter pylori vacA, cagA and iceA genotypes in Nigerian patients with duodenal ulcer disease. J. Med. Microbiol. 2002;51:851–854. - PubMed
-
- Matsuhisa T.M., Yamada N.Y., Kato S.K., Matsukura N.M. Helicobacter pylori infection, mucosal atrophy and intestinal metaplasia in Asian populations: A comparative study in age-, gender- and endoscopic diagnosis-matched subjects. Helicobacter. 2003;8:29–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

