Genomic distribution and inter-sample variation of non-CpG methylation across human cell types
- PMID: 22174693
- PMCID: PMC3234221
- DOI: 10.1371/journal.pgen.1002389
Genomic distribution and inter-sample variation of non-CpG methylation across human cell types
Abstract
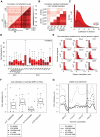
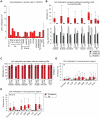
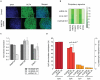
DNA methylation plays an important role in development and disease. The primary sites of DNA methylation in vertebrates are cytosines in the CpG dinucleotide context, which account for roughly three quarters of the total DNA methylation content in human and mouse cells. While the genomic distribution, inter-individual stability, and functional role of CpG methylation are reasonably well understood, little is known about DNA methylation targeting CpA, CpT, and CpC (non-CpG) dinucleotides. Here we report a comprehensive analysis of non-CpG methylation in 76 genome-scale DNA methylation maps across pluripotent and differentiated human cell types. We confirm non-CpG methylation to be predominantly present in pluripotent cell types and observe a decrease upon differentiation and near complete absence in various somatic cell types. Although no function has been assigned to it in pluripotency, our data highlight that non-CpG methylation patterns reappear upon iPS cell reprogramming. Intriguingly, the patterns are highly variable and show little conservation between different pluripotent cell lines. We find a strong correlation of non-CpG methylation and DNMT3 expression levels while showing statistical independence of non-CpG methylation from pluripotency associated gene expression. In line with these findings, we show that knockdown of DNMTA and DNMT3B in hESCs results in a global reduction of non-CpG methylation. Finally, non-CpG methylation appears to be spatially correlated with CpG methylation. In summary these results contribute further to our understanding of cytosine methylation patterns in human cells using a large representative sample set.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Comment in
-
Epigenetics: Dynamic DNA methylation.Nat Rev Genet. 2012 Jan 4;13(2):75. doi: 10.1038/nrg3156. Nat Rev Genet. 2012. PMID: 22215132 No abstract available.
References
-
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. - PubMed
-
- Haines TR, Rodenhiser DI, Ainsworth PJ. Allele-specific non-CpG methylation of the Nf1 gene during early mouse development. Dev Biol. 2001;240:585–598. - PubMed
-
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

