Beyond bacteria: a study of the enteric microbial consortium in extremely low birth weight infants
- PMID: 22174751
- PMCID: PMC3234235
- DOI: 10.1371/journal.pone.0027858
Beyond bacteria: a study of the enteric microbial consortium in extremely low birth weight infants
Abstract
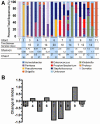
Extremely low birth weight (ELBW) infants have high morbidity and mortality, frequently due to invasive infections from bacteria, fungi, and viruses. The microbial communities present in the gastrointestinal tracts of preterm infants may serve as a reservoir for invasive organisms and remain poorly characterized. We used deep pyrosequencing to examine the gut-associated microbiome of 11 ELBW infants in the first postnatal month, with a first time determination of the eukaryote microbiota such as fungi and nematodes, including bacteria and viruses that have not been previously described. Among the fungi observed, Candida sp. and Clavispora sp. dominated the sequences, but a range of environmental molds were also observed. Surprisingly, seventy-one percent of the infant fecal samples tested contained ribosomal sequences corresponding to the parasitic organism Trichinella. Ribosomal DNA sequences for the roundworm symbiont Xenorhabdus accompanied these sequences in the infant with the greatest proportion of Trichinella sequences. When examining ribosomal DNA sequences in aggregate, Enterobacteriales, Pseudomonas, Staphylococcus, and Enterococcus were the most abundant bacterial taxa in a low diversity bacterial community (mean Shannon-Weaver Index of 1.02 ± 0.69), with relatively little change within individual infants through time. To supplement the ribosomal sequence data, shotgun sequencing was performed on DNA from multiple displacement amplification (MDA) of total fecal genomic DNA from two infants. In addition to the organisms mentioned previously, the metagenome also revealed sequences for gram positive and gram negative bacteriophages, as well as human adenovirus C. Together, these data reveal surprising eukaryotic and viral microbial diversity in ELBW enteric microbiota dominated bytypes of bacteria known to cause invasive disease in these infants.
Conflict of interest statement
Figures


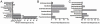
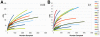
References
-
- Services UDoHaH, editor. US Department of Health and Human Services HRaSA, Maternal and Child Health Bureau. Child Health USA 2010. Rockville, Maryland. 2010.
-
- Davidoff MJ, Dias T, Damus K, Russell R, Bettegowda VR, et al. Changes in the gestational age distribution among U.S. singleton births: impact on rates of late preterm birth, 1992 to 2002. Semin Perinatol. 2006;30:8–15. - PubMed
-
- Wilson-Costello D, Friedman H, Minich N, Fanaroff AA, Hack M. Improved survival rates with increased neurodevelopmental disability for extremely low birth weight infants in the 1990 s. Pediatrics. 2005;115:997–1003. - PubMed
-
- Stoll BJ, Hansen NI, Adams-Chapman I, Fanaroff AA, Hintz SR, et al. Neurodevelopmental and growth impairment among extremely low-birth-weight infants with neonatal infection. Jama. 2004;292:2357–2365. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

