Plantagora: modeling whole genome sequencing and assembly of plant genomes
- PMID: 22174807
- PMCID: PMC3236183
- DOI: 10.1371/journal.pone.0028436
Plantagora: modeling whole genome sequencing and assembly of plant genomes
Abstract
Background: Genomics studies are being revolutionized by the next generation sequencing technologies, which have made whole genome sequencing much more accessible to the average researcher. Whole genome sequencing with the new technologies is a developing art that, despite the large volumes of data that can be produced, may still fail to provide a clear and thorough map of a genome. The Plantagora project was conceived to address specifically the gap between having the technical tools for genome sequencing and knowing precisely the best way to use them.
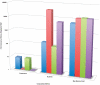
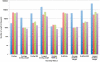
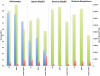
Methodology/principal findings: For Plantagora, a platform was created for generating simulated reads from several different plant genomes of different sizes. The resulting read files mimicked either 454 or Illumina reads, with varying paired end spacing. Thousands of datasets of reads were created, most derived from our primary model genome, rice chromosome one. All reads were assembled with different software assemblers, including Newbler, Abyss, and SOAPdenovo, and the resulting assemblies were evaluated by an extensive battery of metrics chosen for these studies. The metrics included both statistics of the assembly sequences and fidelity-related measures derived by alignment of the assemblies to the original genome source for the reads. The results were presented in a website, which includes a data graphing tool, all created to help the user compare rapidly the feasibility and effectiveness of different sequencing and assembly strategies prior to testing an approach in the lab. Some of our own conclusions regarding the different strategies were also recorded on the website.
Conclusions/significance: Plantagora provides a substantial body of information for comparing different approaches to sequencing a plant genome, and some conclusions regarding some of the specific approaches. Plantagora also provides a platform of metrics and tools for studying the process of sequencing and assembly further.
Conflict of interest statement
Figures

References
-
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The Sequence of the Human Genome. Science. 2001;291:1304–1351. - PubMed
-
- Dib C, Faure S, Fizames C, Samson D, Drouot N, et al. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–154. - PubMed
-
- Settles AM, Byrne M. Opportunities and challenges grow from Arabidopsis genome sequencing. Genome Res. 1998;8:83–85. - PubMed
-
- Bevan M, Mayer K, White O, Eisen JA, Preuss D, et al. Sequence and analysis of the Arabidopsis genome. Current Opinion in Plant Biology. 2001;4:105–110. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

