Regulation of human formyl peptide receptor 1 synthesis: role of single nucleotide polymorphisms, transcription factors, and inflammatory mediators
- PMID: 22174875
- PMCID: PMC3235167
- DOI: 10.1371/journal.pone.0028712
Regulation of human formyl peptide receptor 1 synthesis: role of single nucleotide polymorphisms, transcription factors, and inflammatory mediators
Abstract
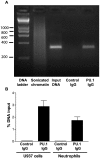
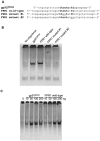
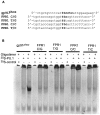
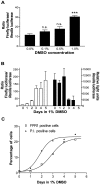
The gene encoding the human formyl peptide receptor 1 (FPR1) is heterogeneous, containing numerous single nucleotide polymorphisms (SNPs). Here, we examine the effect of these SNPs on gene transcription and protein translation. We also identify gene promoter sequences and putative FPR1 transcription factors. To test the effect of codon bias and codon pair bias on FPR1 expression, four FPR1 genetic variants were expressed in human myeloid U937 cells fused to a reporter gene encoding firefly luciferase. No significant differences in luciferase activity were detected, suggesting that the translational regulation and protein stability of FPR1 are modulated by factors other than the SNP codon bias and the variant amino acid properties. Deletion and mutagenesis analysis of the FPR1 promoter showed that a CCAAT box is not required for gene transcription. A -88/41 promoter construct resulted in the strongest transcriptional activity, whereas a -72/41 construct showed large reduction in activity. The region between -88 and -72 contains a consensus binding site for the transcription factor PU.1. Mutagenesis of this site caused significant reduction in reporter gene expression. The PU.1 binding was confirmed in vivo by chromatin immunoprecipitation, and the binding to nucleotides -84 to -76 (TTCCTATTT) was confirmed in vitro by an electrophoretic mobility shift assay. Thus, similar to many other myeloid genes, FPR1 promoter activity requires PU.1. Two single nucleotide polymorphisms at -56 and -54 did not significantly affect FPR1 gene expression, despite differences in binding of transcription factor IRF1 in vitro. Inflammatory mediators such as interferon-γ, tumor necrosis factor-α, and lipopolysaccharide did not increase FPR1 promoter activity in myeloid cells, whereas differentiation induced by DMSO and retinoic acid enhanced the activity. This implies that the expression of FPR1 in myeloid cells is developmentally regulated, and that the differentiated cells are equipped for immediate response to microbial infections.
Conflict of interest statement
Figures
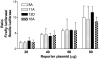





References
-
- Ye RD, Boulay F. Structure and function of leukocyte chemoattractant receptors. Adv Phamacol. 1997;39:221–289. - PubMed
-
- Sahagun-Ruiz A, Colla JS, Juhn J, Gao J-L, Murphy PM, et al. Contrasting evolution of the human leukocyte N-formylpeptide receptor subtypes FPR and FPRL1. Genes Immun. 2001;2:335–342. - PubMed
-
- Zhang Y, Syed R, Uygar C, Pallos D, Gorry MC, et al. Evaluation of human leukocyte N-formyl peptide receptor (FPR1) SNPs in aggressive periodontitis patients. Genes Immun. 2003;4:22–29. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

