Accuracy in copy number calling by qPCR and PRT: a matter of DNA
- PMID: 22174923
- PMCID: PMC3236783
- DOI: 10.1371/journal.pone.0028910
Accuracy in copy number calling by qPCR and PRT: a matter of DNA
Abstract
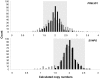
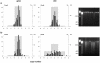
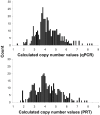
The possible implication of copy number variation (CNV) in the genetic susceptibility to human disease needs to be assessed using robust methods that can be applied at a population scale. In this report, we analyze the performance of the two major techniques, quantitative PCR (qPCR) and paralog ratio test (PRT), and investigate the influence of input DNA amount and template integrity on the reliability of both methods. Analysis of three genes (PRELID1, SYNPO and DEFB4) in a large sample set showed that both methods are prone to false copy number assignments if sufficient attention is not paid to DNA concentration and quality. Accurate normalization of samples is essential for reproducible qPCR because it avoids the effect of differential amplification efficiencies between target and control assays, whereas PRT is generally more sensitive to template degradation due to the fact that longer amplicons are usually needed to optimize sensitivity and specificity of paralog sequence PCR. The use of normalized, high quality genomic DNA yields comparable results with both methods.
Conflict of interest statement
Figures



References
-
- Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37:1243–1246. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

