Autophagy: a primer for the gastroenterologist/hepatologist
- PMID: 22175057
- PMCID: PMC3266158
- DOI: 10.1155/2011/581264
Autophagy: a primer for the gastroenterologist/hepatologist
Abstract
Autophagy is a conserved cellular pathway that maintains intracellular homeostasis by degrading proteins and cytosolic contents of eukaryotic cells. Autophagy clears misfolded and long-lived proteins, damaged organelles and invading microorganisms from cells, and provides nutrients and energy in response to exposure to cell stressors such as starvation. Defective autophagy has recently been linked to a diverse range of disease processes of relevance to gastroenterologists and hepatologists including Crohn's disease, pancreatitis, hepatitis and cancer. The present article provides an overview of the autophagy pathway and discusses gastrointestinal disease processes in which alterations in autophagy have been implicated. The clinical significance of autophagy as a potential therapeutic option is also discussed.
L’autophagie est un mécanisme cellulaire conservé qui maintient l’homéostasie intracellulaire en dégradant les protéines et le contenu cytosolique des cellules eukaryotes. L’autophagie débarrasse la cellule des protéines mal pliées et trop vieilles, des organelles endommagées et des microorganismes envahissants. Elle fournit également à la cellule des nutriments et de l’énergie en réponse à l’exposition aux agresseurs cellulaires, tels que l’inanition. L’autophagie défectueuse a récemment été liée à divers processus pathologiques pertinents pour le gastroentérologue et l’hépatologue, y compris la maladie de Crohn, la pancréatite, l’hépatite et le cancer. Le présent article fournit un aperçu du mécanisme de l’autophagie et expose les processus des maladies gastro-intestinales qui mettent en cause des modifications de l’autophagie. La signification clinique de l’autophagie comme option thérapeutique potentielle est également abordée.
Figures


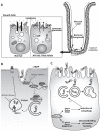


Similar articles
-
Autophagy: healthy eating and self-digestion for gastroenterologists.J Pediatr Gastroenterol Nutr. 2008 May;46(5):496-506. doi: 10.1097/MPG.0b013e3181617895. J Pediatr Gastroenterol Nutr. 2008. PMID: 18493203 Review.
-
An Update Review on the Paneth Cell as Key to Ileal Crohn's Disease.Front Immunol. 2020 Apr 15;11:646. doi: 10.3389/fimmu.2020.00646. eCollection 2020. Front Immunol. 2020. PMID: 32351509 Free PMC article. Review.
-
Autophagy and metabolism.Science. 2010 Dec 3;330(6009):1344-8. doi: 10.1126/science.1193497. Science. 2010. PMID: 21127245 Free PMC article. Review.
-
Crohn's Disease: Potential Drugs for Modulation of Autophagy.Medicina (Kaunas). 2019 May 29;55(6):224. doi: 10.3390/medicina55060224. Medicina (Kaunas). 2019. PMID: 31146413 Free PMC article. Review.
-
Eating the enemy in Crohn's disease: an old theory revisited.J Crohns Colitis. 2010 Oct;4(4):377-83. doi: 10.1016/j.crohns.2010.05.007. Epub 2010 Jun 12. J Crohns Colitis. 2010. PMID: 21122532
Cited by
-
Variants in autophagy genes affect susceptibility to both Crohn's disease and Helicobacter pylori infection.Gastroenterology. 2012 May;142(5):1060-3. doi: 10.1053/j.gastro.2012.03.012. Epub 2012 Mar 23. Gastroenterology. 2012. PMID: 22449582 Free PMC article. No abstract available.
-
New Potential Pharmacological Functions of Chinese Herbal Medicines via Regulation of Autophagy.Molecules. 2016 Mar 17;21(3):359. doi: 10.3390/molecules21030359. Molecules. 2016. PMID: 26999089 Free PMC article. Review.
-
Snapshot: Implications for mTOR in Aging-related Ischemia/Reperfusion Injury.Aging Dis. 2019 Feb 1;10(1):116-133. doi: 10.14336/AD.2018.0501. eCollection 2019 Feb. Aging Dis. 2019. PMID: 30705773 Free PMC article. Review.
-
Hypoxic macrophages impair autophagy in epithelial cells through Wnt1: relevance in IBD.Mucosal Immunol. 2014 Jul;7(4):929-38. doi: 10.1038/mi.2013.108. Epub 2013 Dec 4. Mucosal Immunol. 2014. PMID: 24301659
-
The Role of Heparanase in the Pathogenesis of Acute Pancreatitis: A Potential Therapeutic Target.Sci Rep. 2017 Apr 6;7(1):715. doi: 10.1038/s41598-017-00715-6. Sci Rep. 2017. PMID: 28386074 Free PMC article.
References
-
- de Duve C, Wattiaux R. Functions of lysosomes. Annu Rev Physiol. 1966;28:435–92. - PubMed
-
- Seglen PO, Bohley P. Autophagy and other vacuolar protein degradation mechanisms. Experientia. 1992;48:158–72. - PubMed
-
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–50. - PubMed
-
- Mizushima N, Sugita H, Yoshimori T, Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J Biol Chem. 1998;273:33889–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
