Genetic and phenotypic characterization of sylvatic dengue virus type 4 strains
- PMID: 22178263
- PMCID: PMC3253985
- DOI: 10.1016/j.virol.2011.11.018
Genetic and phenotypic characterization of sylvatic dengue virus type 4 strains
Abstract
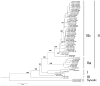
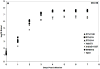
Four serotypes of dengue virus (DENV 1-4) currently circulate between humans and domestic/peridomestic Aedes mosquitoes, resulting in 100 million infections per year. All four serotypes emerged, independently, from sylvatic progenitors transmitted among non-human primates by arboreal Aedes mosquitoes. This study investigated the genetic and phenotypic changes associated with emergence of human DENV-4 from its sylvatic ancestors. Analysis of complete genomes of 3 sylvatic and 4 human strains revealed high conservation of both the 5'- and 3'-untranslated regions but considerable divergence within the open reading frame. Additionally, the two ecotypes did not differ significantly in replication dynamics in cultured human liver (Huh-7), monkey kidney (Vero) or mosquito (C6/36) cells, although significant inter-strain variation within ecotypes was detected. These findings are in partial agreement with previous studies of DENV-2, where human strains produced a larger number of progeny than sylvatic strains in human liver cells but not in monkey or mosquito cells.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures





References
-
- AbuBakar S, Wong PF, Chan YF. Emergence of dengue virus type 4 genotype IIA in Malaysia. J Gen Virol. 2002;83(Pt 10):2437–2442. - PubMed
-
- Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, Gubler DJ, McMillan WO. Selection-driven evolution of emergent dengue virus. Mol Biol Evol. 2003;20(10):1650–1658. - PubMed
-
- Blaney JE, Jr, Hanson CT, Firestone CY, Hanley KA, Murphy BR, Whitehead SS. Genetically modified, live attenuated dengue virus type 3 vaccine candidates. Am J Trop Med Hyg. 2004a;71(6):811–821. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

