Structure, assembly and reading of centromeric chromatin
- PMID: 22178421
- PMCID: PMC3319190
- DOI: 10.1016/j.gde.2011.11.005
Structure, assembly and reading of centromeric chromatin
Abstract
Centromeres are epigenetically defined chromatin domains marked by the presence of the histone H3 variant CENP-A. Here we review recent structural and biochemical work on CENP-A, and advances in understanding the mechanisms that propagate and read centromeric chromatin domains.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

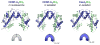

References
-
- Choo KH. Centromerization. Trends Cell Biol. 2000;10:182–188. - PubMed
-
- Maddox PS, Oegema K, Desai A, Cheeseman IM. “Holo”er than thou: chromosome segregation and kinetochore function in C. elegans. Chromosome Res. 2004;12:641–653. - PubMed
-
- Lowell JE, Cross GA. A variant histone H3 is enriched at telomeres in Trypanosoma brucei. J Cell Sci. 2004;117:5937–5947. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

