Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors
- PMID: 22178589
- PMCID: PMC3413201
- DOI: 10.1053/j.gastro.2011.12.005
Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors
Abstract
Background & aims: Cholangiocarcinoma is a heterogeneous disease with a poor outcome that accounts for 5%-10% of primary liver cancers. We characterized its genomic and genetic features and associated these with patient responses to therapy.
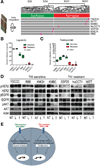
Methods: We profiled the transcriptomes from 104 surgically resected cholangiocarcinoma samples collected from patients in Australia, Europe, and the United States; epithelial and stromal compartments from 23 tumors were laser capture microdissected. We analyzed mutations in KRAS, epidermal growth factor receptor (EGFR), and BRAF in samples from 69 tumors. Changes in gene expression were validated by immunoblotting and immunohistochemistry; integrative genomics combined data from the patients with data from 7 human cholangiocarcinoma cell lines, which were then exposed to trastuzumab and lapatinib.
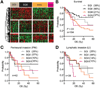
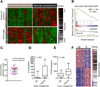
Results: Patients were classified into 2 subclasses, based on 5-year survival rate (72% vs 30%; χ(2) = 11.61; P < .0007), time to recurrence (13.7 vs 22.7 months; P < .001), and the absence or presence of KRAS mutations (24.6%), respectively. Class comparison identified 4 survival subgroups (SGI-IV; χ(2) = 8.34; P < .03); SGIII was characterized by genes associated with proteasomal activity and the worst prognosis. The tumor epithelium was defined by deregulation of the HER2 network and frequent overexpression of EGFR, the hepatocyte growth factor receptor (MET), pRPS6, and Ki67, whereas stroma was enriched in inflammatory cytokines. Lapatinib, an inhibitor of HER2 and EGFR, was more effective in inhibiting growth of cholangiocarcinoma cell lines than trastuzumab.
Conclusions: We provide insight into the pathogenesis of cholangiocarcinoma and identify previously unrecognized subclasses of patients, based on KRAS mutations and increased levels of EGFR and HER2 signaling, who might benefit from dual-target tyrosine kinase inhibitors. The group of patients with the worst prognosis was characterized by transcriptional enrichment of genes that regulate proteasome activity, indicating new therapeutic targets.
Copyright © 2012 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest
The authors disclose no conflicts.
Figures



Comment in
-
Genomic and genetic characterization of cholangiocarcinoma identifies therapeutics targets for tyrosine kinase inhibitors.Gastroenterology. 2012 Oct;143(4):e20-1; author reply e21. doi: 10.1053/j.gastro.2012.07.120. Epub 2012 Aug 22. Gastroenterology. 2012. PMID: 22921673 No abstract available.
References
-
- El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. - PubMed
-
- Deoliveira ML, Schulick RD, Nimura Y, et al. New staging system and a registry for perihilar cholangiocarcinoma. Hepatology. 2011;53:1363–1371. - PubMed
-
- Glimelius B, Hoffman K, Sjoden PO, et al. Chemotherapy improves survival and quality of life in advanced pancreatic and biliary cancer. Ann Oncol. 1996;7:593–600. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

