Metal site occupancy and allosteric switching in bacterial metal sensor proteins
- PMID: 22178748
- PMCID: PMC3312040
- DOI: 10.1016/j.abb.2011.11.021
Metal site occupancy and allosteric switching in bacterial metal sensor proteins
Abstract
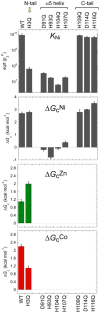
All prokaryotes encode a panel of metal sensor or metalloregulatory proteins that govern the expression of genes that allows an organism to quickly adapt to toxicity or deprivation of both biologically essential transition metal ions, e.g., Zn, Cu, Fe, and heavy metal pollutants. As such, metal sensor proteins can be considered arbiters of intracellular transition metal bioavailability and thus potentially control the metallation state of the metalloproteins in the cell. Metal sensor proteins are specialized allosteric proteins that regulate transcription as a result direct binding of one or two cognate metal ions, to the exclusion of all others. In most cases, the binding of the cognate metal ion induces a structural change in a protein oligomer that either activates or inhibits operator DNA binding. A quantitative measure of the degree to which a particular metal drives metalloregulation of operator DNA-binding is the allosteric coupling free energy, ΔGc. In this review, we summarize recent work directed toward understanding metal occupancy and metal selectivity of these allosteric switches in selected families of metal sensor proteins and examine the structural origins of ΔGc in the functional context a thermodynamic "set-point" model of intracellular metal homeostasis.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures





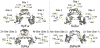
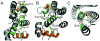



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

