Evolutionary paths to antibiotic resistance under dynamically sustained drug selection
- PMID: 22179135
- PMCID: PMC3534735
- DOI: 10.1038/ng.1034
Evolutionary paths to antibiotic resistance under dynamically sustained drug selection
Abstract
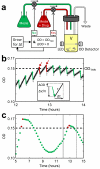
Antibiotic resistance can evolve through the sequential accumulation of multiple mutations. To study such gradual evolution, we developed a selection device, the 'morbidostat', that continuously monitors bacterial growth and dynamically regulates drug concentrations, such that the evolving population is constantly challenged. We analyzed the evolution of resistance in Escherichia coli under selection with single drugs, including chloramphenicol, doxycycline and trimethoprim. Over a period of ∼20 days, resistance levels increased dramatically, with parallel populations showing similar phenotypic trajectories. Whole-genome sequencing of the evolved strains identified mutations both specific to resistance to a particular drug and shared in resistance to multiple drugs. Chloramphenicol and doxycycline resistance evolved smoothly through diverse combinations of mutations in genes involved in translation, transcription and transport. In contrast, trimethoprim resistance evolved in a stepwise manner, through mutations restricted to the gene encoding the enzyme dihydrofolate reductase (DHFR). Sequencing of DHFR over the time course of the experiment showed that parallel populations evolved similar mutations and acquired them in a similar order.
Figures



References
-
- Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312:111–4. - PubMed
-
- Matthews DA, et al. Dihydrofolate reductase: x-ray structure of the binary complex with methotrexate. Science. 1977;197:452–5. - PubMed
-
- Schnell JR, Dyson HJ, Wright PE. Structure, dynamics, and catalytic function of dihydrofolate reductase. Annu Rev Biophys Biomol Struct. 2004;33:119–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

