Highly efficient Staphylococcus carnosus mutant selection system based on suicidal bacteriocin activation
- PMID: 22179253
- PMCID: PMC3273031
- DOI: 10.1128/AEM.06290-11
Highly efficient Staphylococcus carnosus mutant selection system based on suicidal bacteriocin activation
Abstract
Strains from various staphylococcal species produce bacteriocin peptides, which are thought to play important roles in bacterial competition and offer interesting biotechnological avenues. Many bacteriocins are secreted as inactive prepeptides with subsequent activation by specific proteolytic cleavage. By deletion of the protease gene gdmP in Staphylococcus gallinarum Tü3928, which produces the highly active lanthionine-containing bacteriocin gallidermin (lantibiotic), a strain was created producing inactive pregallidermin. On this basis, a new suicidal mutant selection system in the food-grade bacterium Staphylococcus carnosus was developed. Whereas pregallidermin was inactive against S. carnosus, it exerted potent bactericidal activity toward GdmP-secreting S. carnosus strains. To take advantage of this effect, gdmP was cloned in plasmid vectors used for random transposon mutagenesis or targeted allelic replacement of chromosomal genes. Both mutagenesis strategies rely on rare recombination events, and it has remained difficult and laborious to identify mutants among a vast majority of bacterial clones that still contain the delivery vectors. The gdmP-expressing plasmids pGS1 and pGS2 enabled very fast, easy, and reliable identification of transposon and gene replacement mutants, respectively. Mutant selection in the presence of pregallidermin caused suicidal inactivation of all clones that had retained the plasmids and allowed growth of only plasmid-cured mutants. Efficiency of mutant identification was several magnitudes higher than standard screening for the absence of plasmid-encoded antibiotic resistance markers and reached 100% specificity. Thus, the new pregallidermin-based mutant selection system represents a substantial improvement of staphylococcal mutagenesis methodology.
Figures
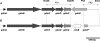



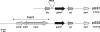

References
-
- Augustin J, Gotz F. 1990. Transformation of Staphylococcus epidermidis and other staphylococcal species with plasmid DNA by electroporation. FEMS Microbiol. Lett. 54:203–207 - PubMed
-
- Augustin J, et al. 1992. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur. J. Biochem. 204:1149–1154 - PubMed
-
- Bae T, Schneewind O. 2006. Allelic replacement in Staphylococcus aureus with inducible counterselection. Plasmid 55:58–63 - PubMed
-
- Bierbaum G, et al. 1996. The biosynthesis of the lantibiotics epidermin, gallidermin, Pep5 and epilancin K7. Antonie Van Leeuwenhoek 69:119–127 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

