Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins
- PMID: 22179551
- PMCID: PMC4165888
- DOI: 10.1038/nmeth.1808
Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins
Abstract
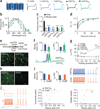
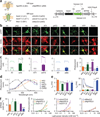
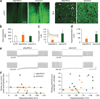
Diverse optogenetic tools have allowed versatile control over neural activity. Many depolarizing and hyperpolarizing tools have now been developed in multiple laboratories and tested across different preparations, presenting opportunities but also making it difficult to draw direct comparisons. This challenge has been compounded by the dependence of performance on parameters such as vector, promoter, expression time, illumination, cell type and many other variables. As a result, it has become increasingly complicated for end users to select the optimal reagents for their experimental needs. For a rapidly growing field, critical figures of merit should be formalized both to establish a framework for further development and so that end users can readily understand how these standardized parameters translate into performance. Here we systematically compared microbial opsins under matched experimental conditions to extract essential principles and identify key parameters for the conduct, design and interpretation of experiments involving optogenetic techniques.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

