Structure of the cyanobacterial Magnesium Chelatase H subunit determined by single particle reconstruction and small-angle X-ray scattering
- PMID: 22179610
- PMCID: PMC3281664
- DOI: 10.1074/jbc.M111.308239
Structure of the cyanobacterial Magnesium Chelatase H subunit determined by single particle reconstruction and small-angle X-ray scattering
Abstract
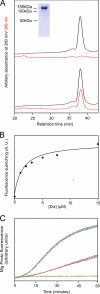
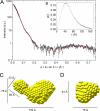
The biosynthesis of chlorophyll, an essential cofactor for photosynthesis, requires the ATP-dependent insertion of Mg(2+) into protoporphyrin IX catalyzed by the multisubunit enzyme magnesium chelatase. This enzyme complex consists of the I subunit, an ATPase that forms a complex with the D subunit, and an H subunit that binds both the protoporphyrin substrate and the magnesium protoporphyrin product. In this study we used electron microscopy and small-angle x-ray scattering to investigate the structure of the magnesium chelatase H subunit, ChlH, from the thermophilic cyanobacterium Thermosynechococcus elongatus. Single particle reconstruction of negatively stained apo-ChlH and Chl-porphyrin proteins was used to reconstitute three-dimensional structures to a resolution of ∼30 Å. ChlH is a large, 148-kDa protein of 1326 residues, forming a cage-like assembly comprising the majority of the structure, attached to a globular N-terminal domain of ∼16 kDa by a narrow linker region. This N-terminal domain is adjacent to a 5 nm-diameter opening in the structure that allows access to a cavity. Small-angle x-ray scattering analysis of ChlH, performed on soluble, catalytically active ChlH, verifies the presence of two domains and their relative sizes. Our results provide a basis for the multiple regulatory and catalytic functions of ChlH of oxygenic photosynthetic organisms and for a chaperoning function that sequesters the enzyme-bound magnesium protoporphyrin product prior to its delivery to the next enzyme in the chlorophyll biosynthetic pathway, magnesium protoporphyrin methyltransferase.
Figures

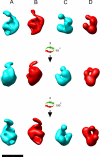

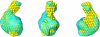
Similar articles
-
Structural and functional consequences of removing the N-terminal domain from the magnesium chelatase ChlH subunit of Thermosynechococcus elongatus.Biochem J. 2014 Dec 15;464(3):315-22. doi: 10.1042/BJ20140463. Biochem J. 2014. PMID: 25471602 Free PMC article.
-
Characterization of the magnesium chelatase from Thermosynechococcus elongatus.Biochem J. 2014 Jan 1;457(1):163-70. doi: 10.1042/BJ20130834. Biochem J. 2014. PMID: 24138165
-
Modification of cysteine residues in the ChlI and ChlH subunits of magnesium chelatase results in enzyme inactivation.Biochem J. 2000 Dec 1;352 Pt 2(Pt 2):435-41. Biochem J. 2000. PMID: 11085937 Free PMC article.
-
Current understanding of the function of magnesium chelatase.Biochem Soc Trans. 2002 Aug;30(4):643-5. doi: 10.1042/bst0300643. Biochem Soc Trans. 2002. PMID: 12196154 Review.
-
Mechanism and regulation of Mg-chelatase.Biochem J. 1997 Oct 15;327 ( Pt 2)(Pt 2):321-33. doi: 10.1042/bj3270321. Biochem J. 1997. PMID: 9359397 Free PMC article. Review.
Cited by
-
Transcriptomic and Structural Insights into Leaf Variegation Development in Ilex × 'Solar Flare'.Int J Mol Sci. 2025 Apr 23;26(9):3999. doi: 10.3390/ijms26093999. Int J Mol Sci. 2025. PMID: 40362242 Free PMC article.
-
Structural insights into the catalytic mechanism of Synechocystis magnesium protoporphyrin IX O-methyltransferase (ChlM).J Biol Chem. 2014 Sep 12;289(37):25690-8. doi: 10.1074/jbc.M114.584920. Epub 2014 Jul 30. J Biol Chem. 2014. PMID: 25077963 Free PMC article.
-
Heterologous Expression of the Barley (Hordeum vulgare L.) Xantha-f, -g and -h Genes that Encode Magnesium Chelatase Subunits.Protein J. 2020 Oct;39(5):554-562. doi: 10.1007/s10930-020-09913-0. Protein J. 2020. PMID: 32737834 Free PMC article.
-
Inhibition of bacteriochlorophyll biosynthesis in the purple phototrophic bacteria Rhodospirillumrubrum and Rhodobacter capsulatus grown in the presence of a toxic concentration of selenite.BMC Microbiol. 2018 Jul 31;18(1):81. doi: 10.1186/s12866-018-1209-5. BMC Microbiol. 2018. PMID: 30064359 Free PMC article.
-
Nanomechanical and Thermophoretic Analyses of the Nucleotide-Dependent Interactions between the AAA(+) Subunits of Magnesium Chelatase.J Am Chem Soc. 2016 May 25;138(20):6591-7. doi: 10.1021/jacs.6b02827. Epub 2016 May 12. J Am Chem Soc. 2016. PMID: 27133226 Free PMC article.
References
-
- Björn L. O., Papageorgiou G. C., Blankenship R. E., Govindjee (2009) A viewpoint. Why chlorophyll a? Photosynth. Res. 99, 85–98 - PubMed
-
- Gibson L. C., Willows R. D., Kannangara C. G., von Wettstein D., Hunter C. N. (1995) Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides. Reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 92, 1941–1944 - PMC - PubMed
-
- Jensen P. E., Gibson L. C., Henningsen K. W., Hunter C. N. (1996) Expression of the chlI, chlD, and chlH genes from the cyanobacterium Synechocystis PCC6803 in Escherichia coli and demonstration that the three cognate proteins are required for magnesium-protoporphyrin chelatase activity. J. Biol. Chem. 271, 16662–16667 - PubMed
-
- Petersen B. L., Jensen P. E., Gibson L. C., Stummann B. M., Hunter C. N., Henningsen K. W. (1998) Reconstitution of an active magnesium chelatase enzyme complex from the bchI, -D, and -H gene products of the green sulfur bacterium Chlorobium vibrioforme expressed in Escherichia coli. J. Bacteriol. 180, 699–704 - PMC - PubMed
-
- Papenbrock J., Gräfe S., Kruse E., Hänel F., Grimm B. (1997) Mg chelatase of tobacco. Identification of a Chl D cDNA sequence encoding a third subunit, analysis of the interaction of the three subunits with the yeast two-hybrid system, and reconstitution of the enzyme activity by co-expression of recombinant CHL D, CHL H and CHL I. Plant J. 12, 981–990 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

