Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA
- PMID: 22179700
- PMCID: PMC3261568
- DOI: 10.1038/emboj.2011.445
Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA
Abstract
Transcription regulation by steroid hormones and other metabolites is mediated by nuclear receptors (NRs) such as the vitamin D and retinoid X receptors (VDR and RXR). Here, we present the cryo electron microscopy (cryo-EM) structure of the heterodimeric complex of the liganded human RXR and VDR bound to a consensus DNA response element forming a direct repeat (DR3). The cryo-EM map of the 100-kDa complex allows positioning the individual crystal structures of ligand- and DNA-binding domains (LBDs and DBDs). The LBDs are arranged perpendicular to the DNA and are located asymmetrically at the DNA 5'-end of the response element. The structure reveals that the VDR N-terminal A/B domain is located close to the DNA. The hinges of both VDR and RXR are fully visible and hold the complex in an open conformation in which co-regulators can bind. The asymmetric topology of the complex provides the structural basis for RXR being an adaptive partner within NR heterodimers, while the specific helical structure of VDR's hinge connects the 3'-bound DBD with the 5'-bound LBD and thereby serves as a conserved linker of defined length sensitive to mutational deletion.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
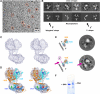
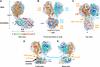
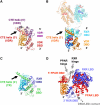
Comment in
-
The nuclear receptor signalling scaffold: insights from full-length structures.EMBO J. 2012 Jan 18;31(2):251-3. doi: 10.1038/emboj.2011.475. Epub 2012 Jan 18. EMBO J. 2012. PMID: 22252143 Free PMC article.
References
-
- Antony P, Sigüeiro R, Huet T, Sato Y, Ramalanjaona N, Rodrigues LC, Mouriño A, Moras D, Rochel N. (2010) Structure-function relationships and crystal structures of the vitamin D receptor bound 2 alpha-methyl-(20S,23S)- and 2 alpha-methyl-(20S,23R)-epoxymethano-1 alpha,25-dihydroxyvitamin D3. J Med Chem 53: 1159–1171 - PubMed
-
- Banerjee P, Chatterjee M (2003) Antiproliferative role of vitamin D and its analogs - a brief overview. Mol Cell Biochem 253: 247–254 - PubMed
-
- Bouillon R, Eelen G, Verlinden L, Mathieu C, Carmeliet G, Verstuyf A. (2006) Vitamin D and cancer. J Steroid Biochem Mol Biol 102: 156–162 - PubMed
-
- Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D (2000) Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell 5: 289–298 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

