Signal-dependent dynamics of transcription factor translocation controls gene expression
- PMID: 22179789
- PMCID: PMC3936303
- DOI: 10.1038/nsmb.2192
Signal-dependent dynamics of transcription factor translocation controls gene expression
Abstract
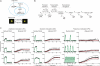
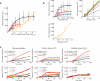
Information about environmental stimuli is often transmitted using common signaling molecules, but the mechanisms that ensure signaling specificity are not entirely known. Here we show that the identities and intensities of different stresses are transmitted by modulation of the amplitude, duration or frequency of nuclear translocation of the Saccharomyces cerevisiae general stress response transcription factor Msn2. Through artificial control of the dynamics of Msn2 translocation, we reveal how distinct dynamical schemes differentially affect reporter gene expression. Using a simple model, we predict stress-induced reporter gene expression from single-cell translocation dynamics. We then demonstrate that the response of natural target genes to dynamical modulation of Msn2 translocation is influenced by differences in the kinetics of promoter transitions and transcription factor binding properties. Thus, multiple environmental signals can trigger qualitatively different dynamics of a single transcription factor and influence gene expression patterns.
Figures





References
-
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–61. - PubMed
-
- Skalhegg BS, Tasken K. Specificity in the cAMP/PKA signaling pathway. Differential expression,regulation, and subcellular localization of subunits of PKA. Front Biosci. 2000;5:D678–93. - PubMed
-
- Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–5. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

