Inhibition of cell migration by PITENINs: the role of ARF6
- PMID: 22179837
- PMCID: PMC3310973
- DOI: 10.1038/onc.2011.593
Inhibition of cell migration by PITENINs: the role of ARF6
Abstract
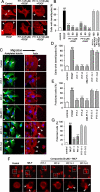
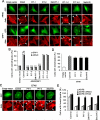
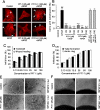
We have reported previously the development of small-molecule phosphatidylinositol-3,4,5-trisphosphate (PIP3) antagonists (PITs) that block pleckstrin homology (PH) domain interaction, including activation of Akt, and show anti-tumor potential. Here we show that the same molecules inhibit growth factor-induced actin remodeling, lamellipodia formation and, ultimately, cell migration and invasion, consistent with an important role of PIP3 in these processes. In vivo, a PIT-1 analog displays significant inhibition on tumor angiogenesis and metastasis. ADP ribosylation factor 6 (ARF6) was recently identified as an important mediator of cytoskeleton and cell motility, which is regulated by PIP3-dependent membrane translocation of the guanine nucleotide exchange factors (GEFs), such as ADP-ribosylation factor nucleotide binding site opener (ARNO) and general receptor for 3-phosphoinositides (GRP1). We demonstrate that PITs inhibit PIP3/ARNO or GRP1 PH domain binding and membrane localization, resulting in the inhibition of ARF6 activation. Importantly, we show that expression of the constitutively active mutant of ARF6 attenuates inhibition of lamellipodia formation and cell migration by PITs, confirming that inhibition of ARF6 contributes to inhibition of these processes by PITs. Overall, our studies demonstrate the feasibility of developing specific small-molecule targeting PIP3 binding by PH domains as potential anticancer agents that can simultaneously interfere with cancer development at multiple points.
Figures





References
-
- Balana ME, Niedergang F, Subtil A, Alcover A, Chavrier P, Dautry-Varsat A. ARF6 GTPase controls bacterial invasion by actin remodelling. J Cell Sci. 2005;118:2201–10. - PubMed
-
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. - PubMed
-
- Caumont AS, Vitale N, Gensse M, Galas MC, Casanova JE, Bader MF. Identification of a plasma membrane-associated guanine nucleotide exchange factor for ARF6 in chromaffin cells. Possible role in the regulated exocytotic pathway. J Biol Chem. 2000;275:15637–44. - PubMed
-
- Cavenagh MM, Whitney JA, Carroll K, Zhang C, Boman AL, Rosenwald AG, et al. Intracellular distribution of Arf proteins in mammalian cells. Arf6 is uniquely localized to the plasma membrane. J Biol Chem. 1996;271:21767–74. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

