Utp23p is required for dissociation of snR30 small nucleolar RNP from preribosomal particles
- PMID: 22180534
- PMCID: PMC3333846
- DOI: 10.1093/nar/gkr1213
Utp23p is required for dissociation of snR30 small nucleolar RNP from preribosomal particles
Abstract
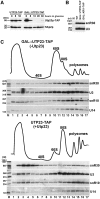
Yeast snR30 is an essential box H/ACA small nucleolar RNA (snoRNA) that promotes 18S rRNA processing through forming transient base-pairing interactions with the newly synthesized 35S pre-rRNA. By using a novel tandem RNA affinity selection approach, followed by coimmunoprecipitation and in vivo cross-linking experiments, we demonstrate that in addition to the four H/ACA core proteins, Cbf5p, Nhp2p, Nop10p and Gar1p, a fraction of snR30 specifically associates with the Utp23p and Kri1p nucleolar proteins. Depletion of Utp23p and Kri1p has no effect on the accumulation and recruitment of snR30 to the nascent pre-ribosomes. However, in the absence of Utp23p, the majority of snR30 accumulates in large pre-ribosomal particles. The retained snR30 is not base-paired with the 35S pre-rRNA, indicating that its aberrant tethering to nascent preribosomes is likely mediated by pre-ribosomal protein(s). Thus, Utp23p may promote conformational changes of the pre-ribosome, essential for snR30 release. Neither Utp23p nor Kri1p is required for recruitment of snR30 to the nascent pre-ribosome. On the contrary, depletion of snR30 prevents proper incorporation of both Utp23p and Kri1p into the 90S pre-ribosome containing the 35S pre-rRNA, indicating that snR30 plays a central role in the assembly of functionally active small subunit processome.
Figures



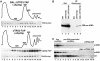

Similar articles
-
Synergistic interaction network between the snR30 RNP, Utp23, and ribosomal RNA during ribosome synthesis.RNA Biol. 2022 Jan;19(1):764-773. doi: 10.1080/15476286.2022.2078092. RNA Biol. 2022. PMID: 35648701 Free PMC article.
-
Identification of novel proteins associated with yeast snR30 small nucleolar RNA.Nucleic Acids Res. 2011 Dec;39(22):9659-70. doi: 10.1093/nar/gkr659. Epub 2011 Sep 5. Nucleic Acids Res. 2011. PMID: 21893585 Free PMC article.
-
H/ACA snR30 snoRNP guides independent 18S rRNA subdomain formation.Nat Commun. 2025 May 21;16(1):4720. doi: 10.1038/s41467-025-59656-8. Nat Commun. 2025. PMID: 40399280 Free PMC article.
-
snR30/U17 Small Nucleolar Ribonucleoprotein: A Critical Player during Ribosome Biogenesis.Cells. 2020 Sep 29;9(10):2195. doi: 10.3390/cells9102195. Cells. 2020. PMID: 33003357 Free PMC article. Review.
-
Sno-capped: 5' ends of preribosomal RNAs are decorated with a U3 SnoRNP.Chem Biol. 2002 Jul;9(7):777-9. doi: 10.1016/s1074-5521(02)00171-0. Chem Biol. 2002. PMID: 12144919 Review.
Cited by
-
Synergistic interaction network between the snR30 RNP, Utp23, and ribosomal RNA during ribosome synthesis.RNA Biol. 2022 Jan;19(1):764-773. doi: 10.1080/15476286.2022.2078092. RNA Biol. 2022. PMID: 35648701 Free PMC article.
-
Structural and functional analysis of Utp24, an endonuclease for processing 18S ribosomal RNA.PLoS One. 2018 Apr 11;13(4):e0195723. doi: 10.1371/journal.pone.0195723. eCollection 2018. PLoS One. 2018. PMID: 29641590 Free PMC article.
-
Structural and functional analysis of Utp23, a yeast ribosome synthesis factor with degenerate PIN domain.RNA. 2013 Dec;19(12):1815-24. doi: 10.1261/rna.040808.113. Epub 2013 Oct 23. RNA. 2013. PMID: 24152547 Free PMC article.
-
The DEAD-box Protein Rok1 Orchestrates 40S and 60S Ribosome Assembly by Promoting the Release of Rrp5 from Pre-40S Ribosomes to Allow for 60S Maturation.PLoS Biol. 2016 Jun 9;14(6):e1002480. doi: 10.1371/journal.pbio.1002480. eCollection 2016 Jun. PLoS Biol. 2016. PMID: 27280440 Free PMC article.
-
Synthetic negative genome screen of the GPN-loop GTPase NPA3 in Saccharomyces cerevisiae.Curr Genet. 2022 Aug;68(3-4):343-360. doi: 10.1007/s00294-022-01243-1. Epub 2022 Jun 4. Curr Genet. 2022. PMID: 35660944
References
-
- Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim. Biophys. Acta. 2010;1803:673–683. - PubMed
-
- Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

