Representation of visual scenes by local neuronal populations in layer 2/3 of mouse visual cortex
- PMID: 22180739
- PMCID: PMC3235640
- DOI: 10.3389/fncir.2011.00018
Representation of visual scenes by local neuronal populations in layer 2/3 of mouse visual cortex
Abstract
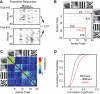
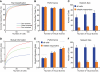
How are visual scenes encoded in local neural networks of visual cortex? In rodents, visual cortex lacks a columnar organization so that processing of diverse features from a spot in visual space could be performed locally by populations of neighboring neurons. To examine how complex visual scenes are represented by local microcircuits in mouse visual cortex we measured visually evoked responses of layer 2/3 neuronal populations using 3D two-photon calcium imaging. Both natural and artificial movie scenes (10 seconds duration) evoked distributed and sparsely organized responses in local populations of 70-150 neurons within the sampled volumes. About 50% of neurons showed calcium transients during visual scene presentation, of which about half displayed reliable temporal activation patterns. The majority of the reliably responding neurons were activated primarily by one of the four visual scenes applied. Consequently, single-neurons performed poorly in decoding, which visual scene had been presented. In contrast, high levels of decoding performance (>80%) were reached when considering population responses, requiring about 80 randomly picked cells or 20 reliable responders. Furthermore, reliable responding neurons tended to have neighbors sharing the same stimulus preference. Because of this local redundancy, it was beneficial for efficient scene decoding to read out activity from spatially distributed rather than locally clustered neurons. Our results suggest a population code in layer 2/3 of visual cortex, where the visual environment is dynamically represented in the activation of distinct functional sub-networks.
Keywords: 3D imaging; calcium imaging; natural movies; neocortex; two-photon microscopy.
Figures





References
-
- Ascoli G. A., Alonso-Nanclares L., Anderson S. A., Barrionuevo G., Benavides-Piccione R., Burkhalter A., Buzsáki G., Cauli B., Defelipe J., Fairén A., Feldmeyer D., Fishell G., Fregnac Y., Freund T. F., Gardner D., Gardner E. P., Goldberg J. H., Helmstaedter M., Hestrin S., Karube F., Kisvárday Z. F., Lambolez B., Lewis D. A., Marin O., Markram H., Muñoz A., Packer A., Petersen C. C., Rockland K. S., Rossier J., Rudy B., Somogyi P., Staiger J. F., Tamas G., Thomson A. M., Toledo-Rodriguez M., Wang Y., West D. C., Yuste R. (2008). Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat. Rev. Neurosci. 9, 557–568 10.1038/nrn2402 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

