Structural Basis for the Antiviral Activity of BST-2/Tetherin and Its Viral Antagonism
- PMID: 22180752
- PMCID: PMC3235769
- DOI: 10.3389/fmicb.2011.00250
Structural Basis for the Antiviral Activity of BST-2/Tetherin and Its Viral Antagonism
Abstract
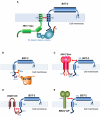
The interferon-inducible host restriction factor bone marrow stromal antigen 2 (BST-2/tetherin) blocks the release of HIV-1 and other enveloped viruses. In turn, these viruses have evolved specific antagonists to counteract this host antiviral molecule, such as the HIV-1 protein Vpu. BST-2 is a type II transmembrane protein with an unusual topology consisting of an N-terminal cytoplasmic tail (CT) followed by a single transmembrane (TM) domain, a coiled-coil extracellular (EC) domain, and a glycosylphosphatidylinositol (GPI) anchor at the C terminus. We and others showed that BST-2 restricts enveloped virus release by bridging the host and virion membranes with its two opposing membrane anchors and that deletion of either one completely abrogates antiviral activity. The EC domain also shows conserved structural properties that are required for antiviral function. It contains several destabilizing amino acids that confer the molecule with conformational flexibility to sustain the protein's function as a virion tether, and three conserved cysteine residues that mediate homodimerization of BST-2, as well as acting as a molecular ruler that separates the membrane anchors. Conversely, the efficient release of virions is promoted by the HIV-1 Vpu protein and other viral antagonists. Our group and others provided evidence from mutational analyses indicating that Vpu antagonism of BST-2-mediated viral restriction requires a highly specific interaction of their mutual TM domains. This interpretation is further supported and expanded by the findings of the latest structural modeling studies showing that critical amino acids in a conserved helical face of these TM domains are required for Vpu-BST-2 interaction and antagonism. In this review, we summarize the current advances in our understanding of the structural basis for BST-2 antiviral function as well as BST-2-specific viral antagonism.
Keywords: BST-2; HIV-1; Vpu; antagonist; interaction; restriction factor; transmembrane.
Figures

References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

