Complete genome sequence of "Enterobacter lignolyticus" SCF1
- PMID: 22180812
- PMCID: PMC3236048
- DOI: 10.4056/sigs.2104875
Complete genome sequence of "Enterobacter lignolyticus" SCF1
Abstract
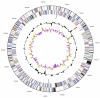
In an effort to discover anaerobic bacteria capable of lignin degradation, we isolated "Enterobacter lignolyticus" SCF1 on minimal media with alkali lignin as the sole source of carbon. This organism was isolated anaerobically from tropical forest soils collected from the Short Cloud Forest site in the El Yunque National Forest in Puerto Rico, USA, part of the Luquillo Long-Term Ecological Research Station. At this site, the soils experience strong fluctuations in redox potential and are net methane producers. Because of its ability to grow on lignin anaerobically, we sequenced the genome. The genome of "E. lignolyticus" SCF1 is 4.81 Mbp with no detected plasmids, and includes a relatively small arsenal of lignocellulolytic carbohydrate active enzymes. Lignin degradation was observed in culture, and the genome revealed two putative laccases, a putative peroxidase, and a complete 4-hydroxyphenylacetate degradation pathway encoded in a single gene cluster.
Keywords: Anaerobic lignin degradation; facultative anaerobe; tropical forest soil isolate.
Figures



Similar articles
-
Complete genome sequence of the lignin-degrading bacterium Klebsiella sp. strain BRL6-2.Stand Genomic Sci. 2014 Dec 8;9:19. doi: 10.1186/1944-3277-9-19. eCollection 2014. Stand Genomic Sci. 2014. PMID: 25566348 Free PMC article.
-
Evidence supporting dissimilatory and assimilatory lignin degradation in Enterobacter lignolyticus SCF1.Front Microbiol. 2013 Sep 19;4:280. doi: 10.3389/fmicb.2013.00280. eCollection 2013. Front Microbiol. 2013. PMID: 24065962 Free PMC article.
-
Multi-time series RNA-seq analysis of Enterobacter lignolyticus SCF1 during growth in lignin-amended medium.PLoS One. 2017 Oct 19;12(10):e0186440. doi: 10.1371/journal.pone.0186440. eCollection 2017. PLoS One. 2017. PMID: 29049419 Free PMC article.
-
Enzyme activities of aerobic lignocellulolytic bacteria isolated from wet tropical forest soils.Syst Appl Microbiol. 2014 Feb;37(1):60-7. doi: 10.1016/j.syapm.2013.10.001. Epub 2013 Nov 14. Syst Appl Microbiol. 2014. PMID: 24238986
-
Upflow anaerobic sludge blanket reactor--a review.Indian J Environ Health. 2001 Apr;43(2):1-82. Indian J Environ Health. 2001. PMID: 12397675 Review.
Cited by
-
Complete genome sequence of the lignin-degrading bacterium Klebsiella sp. strain BRL6-2.Stand Genomic Sci. 2014 Dec 8;9:19. doi: 10.1186/1944-3277-9-19. eCollection 2014. Stand Genomic Sci. 2014. PMID: 25566348 Free PMC article.
-
Characterization of three plant biomass-degrading microbial consortia by metagenomics- and metasecretomics-based approaches.Appl Microbiol Biotechnol. 2016 Dec;100(24):10463-10477. doi: 10.1007/s00253-016-7713-3. Epub 2016 Jul 14. Appl Microbiol Biotechnol. 2016. PMID: 27418359 Free PMC article.
-
Evidence supporting dissimilatory and assimilatory lignin degradation in Enterobacter lignolyticus SCF1.Front Microbiol. 2013 Sep 19;4:280. doi: 10.3389/fmicb.2013.00280. eCollection 2013. Front Microbiol. 2013. PMID: 24065962 Free PMC article.
-
Proteogenomic analysis of a thermophilic bacterial consortium adapted to deconstruct switchgrass.PLoS One. 2013 Jul 19;8(7):e68465. doi: 10.1371/journal.pone.0068465. Print 2013. PLoS One. 2013. PMID: 23894306 Free PMC article.
-
Depolymerization and conversion of lignin to value-added bioproducts by microbial and enzymatic catalysis.Biotechnol Biofuels. 2021 Apr 3;14(1):84. doi: 10.1186/s13068-021-01934-w. Biotechnol Biofuels. 2021. PMID: 33812391 Free PMC article. Review.
References
-
- Cusack DF, Chou WW, Yang WH, Harmon ME, Silver WL. Controls on long-term root and leaf litter decomposition in neotropical forests. Glob Change Biol 2009; 15:1339-1355 10.1111/j.1365-2486.2008.01781.x - DOI
-
- Paul EA, Clark FE. Soil microbiology and biochemistry, Second edition. In: Paul EA, Clark FE, editors. Soil microbiology and biochemistry, Second edition. 1250 Sixth Ave., San Diego, California 92101; 14 Belgrave Square, 24-28 Oval Road, London NW1 70X, England, UK: Academic Press, Inc.; Academic Press Ltd.; 1996. p xiii+340.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

