Accurate proteome-wide protein quantification from high-resolution 15N mass spectra
- PMID: 22182234
- PMCID: PMC3334617
- DOI: 10.1186/gb-2011-12-12-r122
Accurate proteome-wide protein quantification from high-resolution 15N mass spectra
Abstract
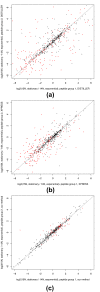
In quantitative mass spectrometry-based proteomics, the metabolic incorporation of a single source of 15N-labeled nitrogen has many advantages over using stable isotope-labeled amino acids. However, the lack of a robust computational framework for analyzing the resulting spectra has impeded wide use of this approach. We have addressed this challenge by introducing a new computational methodology for analyzing 15N spectra in which quantification is integrated with identification. Application of this method to an Escherichia coli growth transition reveals significant improvement in quantification accuracy over previous methods.
Figures