Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents
- PMID: 22183167
- PMCID: PMC3294908
- DOI: 10.1128/AAC.05485-11
Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents
Abstract
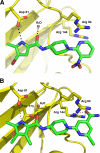
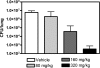
DNA gyrase is an essential enzyme in bacteria, and its inhibition results in the disruption of DNA synthesis and, subsequently, cell death. The pyrrolamides are a novel class of antibacterial agents targeting DNA gyrase. These compounds were identified by a fragment-based lead generation (FBLG) approach using nuclear magnetic resonance (NMR) screening to identify low-molecular-weight compounds that bind to the ATP pocket of DNA gyrase. A pyrrole hit with a binding constant of 1 mM formed the basis of the design and synthesis of a focused library of compounds that resulted in the rapid identification of a lead compound that inhibited DNA gyrase with a 50% inhibitory concentration (IC(50)) of 3 μM. The potency of the lead compound was further optimized by utilizing iterative X-ray crystallography to yield DNA gyrase inhibitors that also displayed antibacterial activity. Spontaneous mutants were isolated in Staphylococcus aureus by plating on agar plates containing pyrrolamide 4 at the MIC. The resistant variants displayed 4- to 8-fold-increased MIC values relative to the parent strain. DNA sequencing revealed two independent point mutations in the pyrrolamide binding region of the gyrB genes from these variants, supporting the hypothesis that the mode of action of these compounds was inhibition of DNA gyrase. Efficacy of a representative pyrrolamide was demonstrated against Streptococcus pneumoniae in a mouse lung infection model. These data demonstrate that the pyrrolamides are a novel class of DNA gyrase inhibitors with the potential to deliver future antibacterial agents targeting multiple clinical indications.
Figures

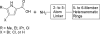
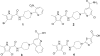
Similar articles
-
Pyrrolamide DNA gyrase inhibitors: optimization of antibacterial activity and efficacy.Bioorg Med Chem Lett. 2011 Dec 15;21(24):7416-20. doi: 10.1016/j.bmcl.2011.10.010. Epub 2011 Oct 12. Bioorg Med Chem Lett. 2011. PMID: 22041057
-
Novel DNA gyrase inhibitors: microbiological characterisation of pyrrolamides.Int J Antimicrob Agents. 2013 Jan;41(1):28-35. doi: 10.1016/j.ijantimicag.2012.08.017. Epub 2012 Nov 8. Int J Antimicrob Agents. 2013. PMID: 23142086
-
Synthesis and Evaluation of N-Phenylpyrrolamides as DNA Gyrase B Inhibitors.ChemMedChem. 2018 Jan 22;13(2):186-198. doi: 10.1002/cmdc.201700549. Epub 2018 Jan 8. ChemMedChem. 2018. PMID: 29206345
-
DNA gyrase inhibitors: Progress and synthesis of potent compounds as antibacterial agents.Biomed Pharmacother. 2018 Jul;103:923-938. doi: 10.1016/j.biopha.2018.04.021. Epub 2018 Apr 24. Biomed Pharmacother. 2018. PMID: 29710509 Review.
-
Recent progress in the discovery and development of DNA gyrase B inhibitors.Future Med Chem. 2018 May 1;10(10):1207-1227. doi: 10.4155/fmc-2017-0257. Epub 2018 May 22. Future Med Chem. 2018. PMID: 29787300 Review.
Cited by
-
A novel high-throughput cell-based assay aimed at identifying inhibitors of DNA metabolism in bacteria.Antimicrob Agents Chemother. 2014 Dec;58(12):7264-72. doi: 10.1128/AAC.03475-14. Epub 2014 Sep 22. Antimicrob Agents Chemother. 2014. PMID: 25246396 Free PMC article.
-
Using Fragment-Based Approaches to Discover New Antibiotics.SLAS Discov. 2018 Jul;23(6):495-510. doi: 10.1177/2472555218773034. SLAS Discov. 2018. PMID: 29923463 Free PMC article.
-
Developments in Non-Intercalating Bacterial Topoisomerase Inhibitors: Allosteric and ATPase Inhibitors of DNA Gyrase and Topoisomerase IV.Pharmaceuticals (Basel). 2023 Feb 8;16(2):261. doi: 10.3390/ph16020261. Pharmaceuticals (Basel). 2023. PMID: 37259406 Free PMC article. Review.
-
Thermodynamic computational approach to capture molecular recognition in the binding of different inhibitors to the DNA gyrase B subunit from Escherichia coli.J Mol Model. 2013 Aug;19(8):3187-200. doi: 10.1007/s00894-013-1849-1. Epub 2013 Apr 30. J Mol Model. 2013. PMID: 23625033
-
Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.ACS Omega. 2020 Apr 24;5(17):10145-10159. doi: 10.1021/acsomega.0c00865. eCollection 2020 May 5. ACS Omega. 2020. PMID: 32391502 Free PMC article.
References
-
- Alanis AJ. 2005. Resistance to antibiotics: Are we in the post-antibiotic era? Arch. Med. Res. 36:697–707 - PubMed
-
- Albert JS, et al. 2007. An integrated approach to fragment-based lead generation: philosophy, strategy and case studies from AstraZeneca's drug discovery programmes. Curr. Top. Med. Chem. 7:1600–1629 - PubMed
-
- Azoulay-Dupuis E, Vallee E, Hardy DJ, Swanson RN, Pocidalo JJ. 1991. Antipneumococcal activity of ciprofloxacin, ofloxacin, and temafloxacin in an experimental mouse pneumonia model at various stages of disease. J. Infect. Dis. 163:319–324 - PubMed
-
- Biedenbach DJ, Farrell DJ, Mendes RE, Ross JE, Jones RN. 2010. Stability of linezolid activity in the era of mobile oxazolidinone resistance determinants: results from the 2009 Zyvox Annuaj Appraisal of Potency and Spectrum program. Diagn. Microbiol. Infect. Dis. 68:459–467 - PubMed
-
- Boucher HW, et al. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48:1–12 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

