Quantitative analysis of genome-wide chromatin remodeling
- PMID: 22183609
- PMCID: PMC6391053
- DOI: 10.1007/978-1-61779-477-3_26
Quantitative analysis of genome-wide chromatin remodeling
Abstract
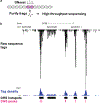
Recent high-throughput sequencing technologies have opened the door for genome-wide characterization of chromatin features at an unprecedented resolution. Chromatin accessibility is an important property that regulates protein binding and other nuclear processes. Here, we describe computational methods to analyze chromatin accessibility using DNaseI hypersensitivity by sequencing (DNaseI-seq). Although there are numerous bioinformatic tools to analyze ChIP-seq data, our statistical algorithm was developed specifically to identify significantly accessible genomic regions by handling features of DNaseI hypersensitivity. Without prior knowledge of relevant protein factors, one can discover genome-wide chromatin remodeling events associated with specific conditions or differentiation stages from quantitative analysis of DNaseI hypersensitivity. By performing appropriate subsequent computational analyses on a select subset of remodeled sites, it is also possible to extract information about putative factors that may bind to specific DNA elements within DNaseI hypersensitive sites. These approaches enabled by DNaseI-seq represent a powerful new methodology that reveals mechanisms of transcriptional regulation.
Figures
References
-
- Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, Weaver M, Shafer A, Lee K, Neri F, Humbert R, Singer MA, Richmond TA, Dorschner MO, McArthur M, Hawrylycz M, Green RD, Navas PA, Noble WS, and Stamatoyannopoulos JA (2006) Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods, 3, 511–518. - PubMed
-
- Hager GL, Nagaich AK, Johnson TA, Walker DA, and John S (2004) Dynamics of nuclear receptor movement and transcription. Biochim. Biophys. Acta, 1677, 46–51. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

