Whole genome sequencing of matched primary and metastatic acral melanomas
- PMID: 22183965
- PMCID: PMC3266028
- DOI: 10.1101/gr.125591.111
Whole genome sequencing of matched primary and metastatic acral melanomas
Abstract
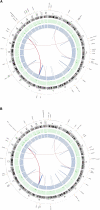
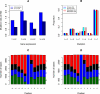
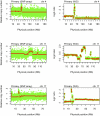
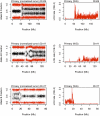
Next generation sequencing has enabled systematic discovery of mutational spectra in cancer samples. Here, we used whole genome sequencing to characterize somatic mutations and structural variation in a primary acral melanoma and its lymph node metastasis. Our data show that the somatic mutational rates in this acral melanoma sample pair were more comparable to the rates reported in cancer genomes not associated with mutagenic exposure than in the genome of a melanoma cell line or the transcriptome of melanoma short-term cultures. Despite the perception that acral skin is sun-protected, the dominant mutational signature in these samples is compatible with damage due to ultraviolet light exposure. A nonsense mutation in ERCC5 discovered in both the primary and metastatic tumors could also have contributed to the mutational signature through accumulation of unrepaired dipyrimidine lesions. However, evidence of transcription-coupled repair was suggested by the lower mutational rate in the transcribed regions and expressed genes. The primary and the metastasis are highly similar at the level of global gene copy number alterations, loss of heterozygosity and single nucleotide variation (SNV). Furthermore, the majority of the SNVs in the primary tumor were propagated in the metastasis and one nonsynonymous coding SNV and one splice site mutation appeared to arise de novo in the metastatic lesion.
Figures
References
-
- Albert SM 2010. Neurodegenerative disease and cancer: A critical role for melanoma? Neuroepidemiology 35: 305–306 - PubMed
-
- Aung CS, Hill MM, Bastiani M, Parton RG, Parat MO 2011. PTRF-cavin-1 expression decreases the migration of PC3 prostate cancer cells: Role of matrix metalloprotease 9. Eur J Cell Biol 90: 136–142 - PubMed
-
- Bastian BC, Kashani-Sabet M, Hamm H, Godfrey T, Moore DH 2nd, Brocker EB, LeBoit PE, Pinkel D 2000. Gene amplifications characterize acral melanoma and permit the detection of occult tumor cells in the surrounding skin. Cancer Res 60: 1968–1973 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
