DNA damage signals through differentially modified E2F1 molecules to induce apoptosis
- PMID: 22184068
- PMCID: PMC3295199
- DOI: 10.1128/MCB.06286-11
DNA damage signals through differentially modified E2F1 molecules to induce apoptosis
Abstract
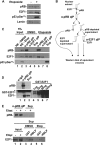
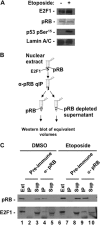
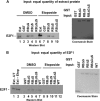
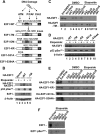
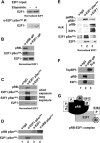
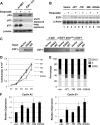
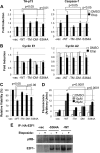
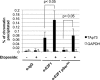
E2F transcription can lead to cell proliferation or apoptosis, indicating that E2Fs control opposing functions. In a similar manner, DNA double-strand breaks can signal to induce cell cycle arrest or apoptosis. Specifically, pRB is activated following DNA damage, allowing it to bind to E2Fs and block transcription at cell cycle promoters; however, E2F1 is simultaneously activated, leading to transcription at proapoptotic promoters. We examined this paradoxical control of E2F transcription by studying how E2F1's interaction with pRB is regulated following DNA damage. Our work reveals that DNA damage signals create multiple forms of E2F1 that contain mutually exclusive posttranslational modifications. Specifically, E2F1 phospho-serine 364 is found only in complex with pRB, while E2F1 phosphorylation at serine 31 and acetylation function to create a pRB-free form of E2F1. Both pRB-bound and pRB-free modifications on E2F1 are essential for the activation of TA-p73 and the maximal induction of apoptosis. Chromatin immunoprecipitation demonstrated that E2F1 phosphorylated on serine 364 is also present at proapoptotic gene promoters during the induction of apoptosis. This indicates that distinct populations of E2F1 are organized in response to DNA damage signaling. Surprisingly, these complexes act in parallel to activate transcription of proapoptotic genes. Our data suggest that DNA damage signals alter pRB and E2F1 to engage them in functions leading to apoptotic induction that are distinct from pRB-E2F regulation in cell cycle control.
Figures
References
-
- Aparicio O, et al. 2005. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo, p 21.23.21–21.23.33 In Ausubel FM, Brent R, Kington RE, Moore DD, Seidman JG, Smith JA, Struhl KE. (ed), Current protocols in molecular biology. Greene Publishing Associates, New York, NY - PubMed
-
- Avni D, et al. 2003. Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol. Cell 12:735–746 - PubMed
-
- Bates S, et al. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124–125 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
