Type 1 diabetes-associated HLA-DQ8 transdimer accommodates a unique peptide repertoire
- PMID: 22184118
- PMCID: PMC3308765
- DOI: 10.1074/jbc.M111.313940
Type 1 diabetes-associated HLA-DQ8 transdimer accommodates a unique peptide repertoire
Abstract
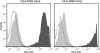
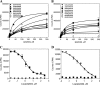
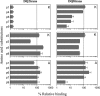
HLA-DQ2 and HLA-DQ8 are strongly predisposing haplotypes for type 1 diabetes (T1D). Yet HLA-DQ2/8 heterozygous individuals have a synergistically increased risk compared with HLA-DQ2 or HLA-DQ8 homozygote subjects that may result from the presence of a transdimer formed between the α-chain of HLA-DQ2 (DQA1*05:01) and the β-chain of HLA-DQ8 (DQB1*03:02). We generated cells exclusively expressing this transdimer (HLA-DQ8trans), characterized its peptide binding repertoire, and defined a unique transdimer-specific peptide binding motif that was found to be distinct from those of HLA-DQ2 and HLA-DQ8. This motif predicts an array of peptides of islet autoantigens as candidate T cell epitopes, many of which selectively bind to the HLA transdimer, whereas others bind to both HLA-DQ8 and transdimer with similar affinity. Our findings provide a molecular basis for the association between HLA-DQ transdimers and T1D and set the stage for rational testing of potential diabetogenic peptide epitopes.
Figures



References
-
- Atkinson M. A., Maclaren N. K. (1994) The pathogenesis of insulin-dependent diabetes mellitus. N. Engl. J. Med. 331, 1428–1436 - PubMed
-
- van Belle T. L., Coppieters K. T., von Herrath M. G. (2011) Type 1 diabetes. Etiology, immunology, and therapeutic strategies. Physiol. Rev. 91, 79–118 - PubMed
-
- Kent S. C., Chen Y., Bregoli L., Clemmings S. M., Kenyon N. S., Ricordi C., Hering B. J., Hafler D. A. (2005) Expanded T cells from pancreatic lymph nodes of type 1 diabetic subjects recognize an insulin epitope. Nature 435, 224–228 - PubMed
-
- Miller G. G., Pollack M. S., Nell L. J., Thomas J. W. (1987) Insulin-specific human T cells. Epitope specificity, major histocompatibility complex restriction, and alloreactivity to a diabetes-associated haplotype. J. Immunol. 139, 3622–3629 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

