Exploring one-state downhill protein folding in single molecules
- PMID: 22184219
- PMCID: PMC3252948
- DOI: 10.1073/pnas.1111164109
Exploring one-state downhill protein folding in single molecules
Abstract
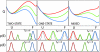
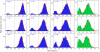
A one-state downhill protein folding process is barrierless at all conditions, resulting in gradual melting of native structure that permits resolving folding mechanisms step-by-step at atomic resolution. Experimental studies of one-state downhill folding have typically focused on the thermal denaturation of proteins that fold near the speed limit (ca. 10(6) s(-1)) at their unfolding temperature, thus being several orders of magnitude too fast for current single-molecule methods, such as single-molecule FRET. An important open question is whether one-state downhill folding kinetics can be slowed down to make them accessible to single-molecule approaches without turning the protein into a conventional activated folder. Here we address this question on the small helical protein BBL, a paradigm of one-state downhill thermal (un)folding. We decreased 200-fold the BBL folding-unfolding rate by combining chemical denaturation and low temperature, and carried out free-diffusion single-molecule FRET experiments with 50-μs resolution and maximal photoprotection using a recently developed Trolox-cysteamine cocktail. These experiments revealed a single conformational ensemble at all denaturing conditions. The chemical unfolding of BBL was then manifested by the gradual change of this unique ensemble, which shifts from high to low FRET efficiency and becomes broader at increasing denaturant. Furthermore, using detailed quantitative analysis, we could rule out the possibility that the BBL single-molecule data are produced by partly overlapping folded and unfolded peaks. Thus, our results demonstrate the one-state downhill folding regime at the single-molecule level and highlight that this folding scenario is not necessarily associated with ultrafast kinetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
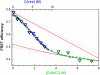


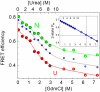
 obtained from the FEH (blue small circles) are also shown for reference. (Inset) Changes in native probability as function of denaturant concentration obtained from the fit.
obtained from the FEH (blue small circles) are also shown for reference. (Inset) Changes in native probability as function of denaturant concentration obtained from the fit.
Comment in
-
Don't waste good methods on bad buffers and ambiguous data.Proc Natl Acad Sci U S A. 2013 Jan 29;110(5):E331-2. doi: 10.1073/pnas.1217840110. Epub 2013 Jan 17. Proc Natl Acad Sci U S A. 2013. PMID: 23329326 Free PMC article. No abstract available.
Similar articles
-
Slow proton transfer coupled to unfolding explains the puzzling results of single-molecule experiments on BBL, a paradigmatic downhill folding protein.PLoS One. 2013 Oct 28;8(10):e78044. doi: 10.1371/journal.pone.0078044. eCollection 2013. PLoS One. 2013. PMID: 24205082 Free PMC article.
-
Direct observation of barrier-limited folding of BBL by single-molecule fluorescence resonance energy transfer.Proc Natl Acad Sci U S A. 2009 Sep 22;106(38):16239-44. doi: 10.1073/pnas.0909126106. Epub 2009 Sep 11. Proc Natl Acad Sci U S A. 2009. PMID: 19805287 Free PMC article.
-
Downhill versus barrier-limited folding of BBL 2: mechanistic insights from kinetics of folding monitored by independent tryptophan probes.J Mol Biol. 2009 Apr 10;387(4):975-85. doi: 10.1016/j.jmb.2008.12.056. Epub 2008 Dec 30. J Mol Biol. 2009. PMID: 19136014
-
The study of protein folding and dynamics by determination of intramolecular distance distributions and their fluctuations using ensemble and single-molecule FRET measurements.Chemphyschem. 2005 May;6(5):858-70. doi: 10.1002/cphc.200400617. Chemphyschem. 2005. PMID: 15884068 Review.
-
Limited cooperativity in protein folding.Curr Opin Struct Biol. 2016 Feb;36:58-66. doi: 10.1016/j.sbi.2015.12.001. Epub 2016 Feb 2. Curr Opin Struct Biol. 2016. PMID: 26845039 Review.
Cited by
-
Reply to Campos and Muñoz: Why phosphate is a bad buffer for guanidinium chloride titrations.Proc Natl Acad Sci U S A. 2013 Apr 2;110(14):E1244-5. doi: 10.1073/pnas.1303286110. Proc Natl Acad Sci U S A. 2013. PMID: 23675589 Free PMC article. No abstract available.
-
Don't waste good methods on bad buffers and ambiguous data.Proc Natl Acad Sci U S A. 2013 Jan 29;110(5):E331-2. doi: 10.1073/pnas.1217840110. Epub 2013 Jan 17. Proc Natl Acad Sci U S A. 2013. PMID: 23329326 Free PMC article. No abstract available.
-
Molten globule-like transition state of protein barnase measured with calorimetric force spectroscopy.Proc Natl Acad Sci U S A. 2022 Mar 15;119(11):e2112382119. doi: 10.1073/pnas.2112382119. Epub 2022 Mar 10. Proc Natl Acad Sci U S A. 2022. PMID: 35271392 Free PMC article.
-
Ultrafast folding kinetics of WW domains reveal how the amino acid sequence determines the speed limit to protein folding.Proc Natl Acad Sci U S A. 2019 Apr 23;116(17):8137-8142. doi: 10.1073/pnas.1900203116. Epub 2019 Apr 9. Proc Natl Acad Sci U S A. 2019. PMID: 30967507 Free PMC article.
-
Equilibrium unfolding of the PDZ domain of β2-syntrophin.Biophys J. 2012 Jun 20;102(12):2835-44. doi: 10.1016/j.bpj.2012.05.021. Epub 2012 Jun 19. Biophys J. 2012. PMID: 22735534 Free PMC article.
References
-
- Bryngelson JD, Onuchic JN, Socci ND, Wolynes PG. Funnels, pathways, and the energy landscape of protein-folding—a synthesis. Proteins Struct Funct Genet. 1995;21:167–195. - PubMed
-
- Bustamante C, Chemla YR, Forde NR, Izhaky D. Mechanical processes in biochemistry. Annu Rev Biochem. 2004;73:705–748. - PubMed
-
- Schuler B, Lipman EA, Eaton WA. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 2002;419:743–747. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

