Geminin overexpression induces mammary tumors via suppressing cytokinesis
- PMID: 22184288
- PMCID: PMC3282064
- DOI: 10.18632/oncotarget.363
Geminin overexpression induces mammary tumors via suppressing cytokinesis
Abstract
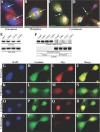
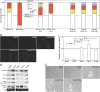
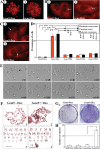
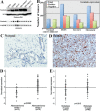
Aneuploidy plays an important role in the development of cancer. Here, we uncovered an oncogenic role for geminin in mitotic cells. In addition to chromatin, tyrosine phosphorylated geminin also localizes to centrosome, spindle, cleavage furrow and midbody during mitosis. Geminin binding to Aurora B prevents its binding to INCENP, and thus activation leading to lack of histone H3-(serine 10) phosphorylation, chromosome condensation failure, aborted cytokinesis and the formation of aneuploid, drug resistance cells. Geminin overexpressing human mammary epithelial cells form aneuploid, aggressive tumors in SCID mice. Geminin is overexpressed in more than half of all breast cancers analyzed. The current study reveals that geminin is a genuine oncogene that promotes cytokinesis failure and production of aneuploid, aggressive breast tumors when overexpressed and thus a worthy therapeutic target (oncotarget) for aggressive breast cancer.
Figures



References
-
- Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol. 2004;5:45–54. - PubMed
-
- Eggert U, Mitchison T, Field C. Animal cytokinesis: from parts list to mechanisms. Ann Rev Biochem. 2006;75:543–566. - PubMed
-
- Ganem N, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. - PubMed
-
- Guizetti J, Gerlich D. Cytokinetic abscission in animal cells. Semin Cell Dev Biol. 2010;21(9):909–916. - PubMed
-
- Cimini D, Wan X, Hirel C, Salmon E. Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol. 2006;16:1711–1718. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

