Use of the chinchilla model for nasopharyngeal colonization to study gene expression by Moraxella catarrhalis
- PMID: 22184412
- PMCID: PMC3294655
- DOI: 10.1128/IAI.05918-11
Use of the chinchilla model for nasopharyngeal colonization to study gene expression by Moraxella catarrhalis
Abstract
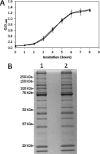
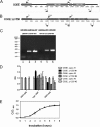
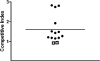
Young adult chinchillas were atraumatically inoculated with Moraxella catarrhalis via the nasal route. Detailed histopathologic examination of nasopharyngeal tissues isolated from these M. catarrhalis-infected animals revealed the presence of significant inflammation within the epithelium. Absence of similar histopathologic findings in sham-inoculated animals confirmed that M. catarrhalis was exposed to significant host-derived factors in this environment. Twenty-four hours after inoculation, viable M. catarrhalis organisms were recovered from the nasal cavity and nasopharynx of the animals in numbers sufficient for DNA microarray analysis. More than 100 M. catarrhalis genes were upregulated in vivo, including open reading frames (ORFs) encoding proteins that are involved in a truncated denitrification pathway or in the oxidative stress response, as well as several putative transcriptional regulators. Additionally, 200 M. catarrhalis genes were found to be downregulated when this bacterium was introduced into the nasopharynx. These downregulated genes included ORFs encoding several well-characterized M. catarrhalis surface proteins including Hag, McaP, and MchA1. Real-time reverse transcriptase PCR (RT-PCR) was utilized as a stringent control to validate the results of in vivo gene expression patterns as measured by DNA microarray analysis. Inactivation of one of the genes (MC ORF 1550) that was upregulated in vivo resulted in a decrease in the ability of M. catarrhalis to survive in the chinchilla nasopharynx over a 3-day period. This is the first evaluation of global transcriptome expression by M. catarrhalis cells in vivo.
Figures

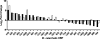
References
Publication types
MeSH terms
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

