Adaptation to tRNA acceptor stem structure by flexible adjustment in the catalytic domain of class I tRNA synthetases
- PMID: 22184460
- PMCID: PMC3264908
- DOI: 10.1261/rna.029983.111
Adaptation to tRNA acceptor stem structure by flexible adjustment in the catalytic domain of class I tRNA synthetases
Abstract
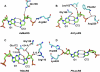
Class I aminoacyl-tRNA synthetases (aaRSs) use a Rossmann-fold domain to catalyze the synthesis of aminoacyl-tRNAs required for decoding genetic information. While the Rossmann-fold domain is conserved in evolution, the acceptor stem near the aminoacylation site varies among tRNA substrates, raising the question of how the conserved protein fold adapts to RNA sequence variations. Of interest is the existence of an unpaired C-A mismatch at the 1-72 position unique to bacterial initiator tRNA(fMet) and absent from elongator tRNAs. Here we show that the class I methionyl-tRNA synthetase (MetRS) of Escherichia coli and its close structural homolog cysteinyl-tRNA synthetase (CysRS) display distinct patterns of recognition of the 1-72 base pair. While the structural homology of the two enzymes in the Rossmann-fold domain is manifested in a common burst feature of aminoacylation kinetics, CysRS discriminates against unpaired 1-72, whereas MetRS lacks such discrimination. A structure-based alignment of the Rossmann fold identifies the insertion of an α-helical motif, specific to CysRS but absent from MetRS, which docks on 1-72 and may discriminate against mismatches. Indeed, substitutions of the CysRS helical motif abolish the discrimination against unpaired 1-72. Additional structural alignments reveal that with the exception of MetRS, class I tRNA synthetases contain a structural motif that docks on 1-72. This work demonstrates that by flexible insertion of a structural motif to dock on 1-72, the catalytic domain of class I tRNA synthetases can acquire structural plasticity to adapt to changes at the end of the tRNA acceptor stem.
Figures


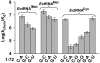
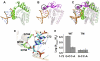
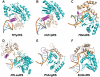
Similar articles
-
Influence of transfer RNA tertiary structure on aminoacylation efficiency by glutaminyl and cysteinyl-tRNA synthetases.J Mol Biol. 2000 Jun 2;299(2):431-46. doi: 10.1006/jmbi.2000.3749. J Mol Biol. 2000. PMID: 10860750
-
The 2.0 A crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules.Structure. 2000 Feb 15;8(2):197-208. doi: 10.1016/s0969-2126(00)00095-2. Structure. 2000. PMID: 10673435
-
Role for a conserved structural motif in assembly of a class I aminoacyl-tRNA synthetase active site.Biochemistry. 2011 Feb 8;50(5):763-9. doi: 10.1021/bi101375d. Epub 2011 Jan 11. Biochemistry. 2011. PMID: 21175197
-
Domain-domain communication in aminoacyl-tRNA synthetases.Prog Nucleic Acid Res Mol Biol. 2001;69:317-49. doi: 10.1016/s0079-6603(01)69050-0. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11550797 Review.
-
Aminoacylation of RNA minihelices: implications for tRNA synthetase structural design and evolution.Crit Rev Biochem Mol Biol. 1993;28(4):309-22. doi: 10.3109/10409239309078438. Crit Rev Biochem Mol Biol. 1993. PMID: 7691478 Review.
Cited by
-
Cys-tRNAj as a Second Translation Initiator for Priming Proteins with Cysteine in Bacteria.ACS Omega. 2025 Jan 29;10(5):4548-4560. doi: 10.1021/acsomega.4c08326. eCollection 2025 Feb 11. ACS Omega. 2025. PMID: 39959092 Free PMC article.
-
Loss-of-function mutations in Lysyl-tRNA synthetase cause various leukoencephalopathy phenotypes.Neurol Genet. 2019 Apr 18;5(2):e565. doi: 10.1212/NXG.0000000000000316. eCollection 2019 Apr. Neurol Genet. 2019. PMID: 31192300 Free PMC article.
-
Transfer RNAs with novel cloverleaf structures.Nucleic Acids Res. 2017 Mar 17;45(5):2776-2785. doi: 10.1093/nar/gkw898. Nucleic Acids Res. 2017. PMID: 28076288 Free PMC article.
-
Kinetic Analysis of tRNA Methyltransferases.Methods Enzymol. 2015;560:91-116. doi: 10.1016/bs.mie.2015.04.012. Epub 2015 Jun 2. Methods Enzymol. 2015. PMID: 26253967 Free PMC article.
-
Molecular Basis and Consequences of the Cytochrome c-tRNA Interaction.J Biol Chem. 2016 May 6;291(19):10426-36. doi: 10.1074/jbc.M115.697789. Epub 2016 Mar 9. J Biol Chem. 2016. PMID: 26961879 Free PMC article.
References
-
- Bullock TL, Uter N, Nissan TA, Perona JJ 2003. Amino acid discrimination by a class I aminoacyl-tRNA synthetase specified by negative determinants. J Mol Biol 328: 395–408 - PubMed
-
- Eldred EW, Schimmel PR 1972. Investigation of the transfer of amino acid from a transfer ribonucleic acid synthetase-aminoacyl adenylate complex to transfer ribonucleic acid. Biochemistry 11: 17–23 - PubMed
-
- Fechter P, Rudinger J, Giege R, Theobald-Dietrich A 1998. Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: production and activity. FEBS Lett 436: 99–103 - PubMed
-
- Fersht AR, Dingwall C 1979. Cysteinyl-tRNA synthetase from Escherichia coli does not need an editing mechanism to reject serine and alanine. High binding energy of small groups in specific molecular interactions. Biochemistry 18: 1245–1249 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
