Genetic and epigenetic variations contributed by Alu retrotransposition
- PMID: 22185517
- PMCID: PMC3272032
- DOI: 10.1186/1471-2164-12-617
Genetic and epigenetic variations contributed by Alu retrotransposition
Abstract
Background: De novo retrotransposition of Alu elements has been recognized as a major driver for insertion polymorphisms in human populations. In this study, we exploited Alu-anchored bisulfite PCR libraries to identify evolutionarily recent Alu element insertions, and to investigate their genetic and epigenetic variation.
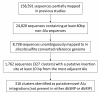
Results: A total of 327 putatively recent Alu insertions were identified, altogether represented by 1,762 sequence reads. Nearly all such de novo retrotransposition events (316/327) were novel. Forty-seven out of forty-nine randomly selected events, corresponding to nineteen genomic loci, were sequence-verified. Alu element insertions remained hemizygous in one or more individuals in sixteen of the nineteen genomic loci. The Alu elements were found to be enriched for young Alu families with characteristic sequence features, such as the presence of a longer poly(A) tail. In addition, we documented the occurrence of a duplication of the AT-rich target site in their immediate flanking sequences, a hallmark of retrotransposition. Furthermore, we found the sequence motif (TT/AAAA) that is recognized by the ORF2P protein encoded by LINE-1 in their 5'-flanking regions, consistent with the fact that Alu retrotransposition is facilitated by LINE-1 elements. While most of these Alu elements were heavily methylated, we identified an Alu localized 1.5 kb downstream of TOMM5 that exhibited a completely unmethylated left arm. Interestingly, we observed differential methylation of its immediate 5' and 3' flanking CpG dinucleotides, in concordance with the unmethylated and methylated statuses of its internal 5' and 3' sequences, respectively. Importantly, TOMM5's CpG island and the 3 Alu repeats and 1 MIR element localized upstream of this newly inserted Alu were also found to be unmethylated. Methylation analyses of two additional genomic loci revealed no methylation differences in CpG dinucleotides flanking the Alu insertion sites in the two homologous chromosomes, irrespective of the presence or absence of the insertion.
Conclusions: We anticipate that the combination of methodologies utilized in this study, which included repeat-anchored bisulfite PCR sequencing and the computational analysis pipeline herein reported, will prove invaluable for the generation of genetic and epigenetic variation maps.
Figures



References
-
- Weiner AM. An abundant cytoplasmic 7 S RNA is complementary to the dominant interspersed middle repetitive DNA sequence family in the human genome. Cell. 1980;22(1 Pt 1):209–218. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

