Clonal and molecular analysis of the prospective anterior neural boundary in the mouse embryo
- PMID: 22186731
- PMCID: PMC3243100
- DOI: 10.1242/dev.075499
Clonal and molecular analysis of the prospective anterior neural boundary in the mouse embryo
Abstract
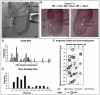
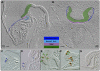
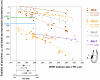
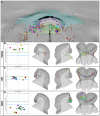
In the mouse embryo the anterior ectoderm undergoes extensive growth and morphogenesis to form the forebrain and cephalic non-neural ectoderm. We traced descendants of single ectoderm cells to study cell fate choice and cell behaviour at late gastrulation. In addition, we provide a comprehensive spatiotemporal atlas of anterior gene expression at stages crucial for anterior ectoderm regionalisation and neural plate formation. Our results show that, at late gastrulation stage, expression patterns of anterior ectoderm genes overlap significantly and correlate with areas of distinct prospective fates but do not define lineages. The fate map delineates a rostral limit to forebrain contribution. However, no early subdivision of the presumptive forebrain territory can be detected. Lineage analysis at single-cell resolution revealed that precursors of the anterior neural ridge (ANR), a signalling centre involved in forebrain development and patterning, are clonally related to neural ectoderm. The prospective ANR and the forebrain neuroectoderm arise from cells scattered within the same broad area of anterior ectoderm. This study establishes that although the segregation between non-neural and neural precursors in the anterior midline ectoderm is not complete at late gastrulation stage, this tissue already harbours elements of regionalisation that prefigure the later organisation of the head.
Figures




References
-
- Acampora D., Merlo G. R., Paleari L., Zerega B., Postiglione M. P., Mantero S., Bober E., Barbieri O., Simeone A., Levi G. (1999). Craniofacial, vestibular and bone defects in mice lacking the Distal-less-related gene Dlx5. Development 126, 3795-3809 - PubMed
-
- Acampora D., Postiglione M. P., Avantaggiato V., Di Bonito M., Simeone A. (2000). The role of Otx and Otp genes in brain development. Int. J. Dev. Biol. 44, 669-677 - PubMed
-
- Ang S. L., Rossant J. (1993). Anterior mesendoderm induces mouse Engrailed genes in explant cultures. Development 118, 139-149 - PubMed
-
- Ang S. L., Conlon R. A., Jin O., Rossant J. (1994). Positive and negative signals from mesoderm regulate the expression of mouse Otx2 in ectoderm explants. Development 120, 2979-2989 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

