5-Hydroxymethylcytosine: a new kid on the epigenetic block?
- PMID: 22186736
- PMCID: PMC3737735
- DOI: 10.1038/msb.2011.95
5-Hydroxymethylcytosine: a new kid on the epigenetic block?
Abstract
The discovery of the Ten-Eleven-Translocation (TET) oxygenases that catalyze the hydroxylation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) has triggered an avalanche of studies aiming to resolve the role of 5hmC in gene regulation if any. Hitherto, TET1 is reported to bind to CpG-island (CGI) and bivalent promoters in mouse embryonic stem cells, whereas binding at DNAseI hypersensitive sites (HS) had escaped previous analysis. Significant enrichment/accumulation of 5hmC but not 5mC can indeed be detected at bivalent promoters and at DNaseI-HS. Surprisingly, however, 5hmC is not detected or present at very low levels at CGI promoters notwithstanding the presence of TET1. Our meta-analysis of DNA methylation profiling points to potential issues with regard to the various methodologies that are part of the toolbox used to detect 5mC and 5hmC. Discrepancies between published studies and technical limitations prevent an unambiguous assignment of 5hmC as a 'true' epigenetic mark, that is, read and interpreted by other factors and/or as a transiently accumulating intermediary product of the conversion of 5mC to unmodified cytosines.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
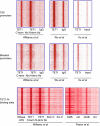

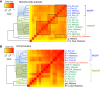


References
-
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 - PubMed
-
- Brinkman AB, Simmer F, Ma K, Kaan A, Zhu J, Stunnenberg HG (2010) Whole-genome DNA methylation profiling using MethylCap-seq. Methods 52: 232–236 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

