Functional regulation of pre-B-cell leukemia homeobox interacting protein 1 (PBXIP1/HPIP) in erythroid differentiation
- PMID: 22187427
- PMCID: PMC3285334
- DOI: 10.1074/jbc.M111.289843
Functional regulation of pre-B-cell leukemia homeobox interacting protein 1 (PBXIP1/HPIP) in erythroid differentiation
Abstract
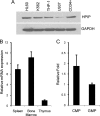
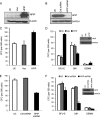
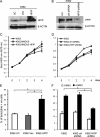
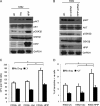
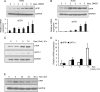
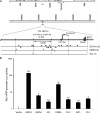
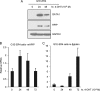
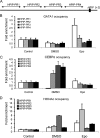
Pre-B-cell leukemia homeobox interacting protein 1 or human PBX1 interacting protein (PBXIP1/HPIP) is a co-repressor of pre-B-cell leukemia homeobox 1 (PBX1) and is also known to regulate estrogen receptor functions by associating with the microtubule network. Despite its initial discovery in the context of hematopoietic cells, little is yet known about the role of HPIP in hematopoiesis. Here, we show that lentivirus-mediated overexpression of HPIP in human CD34(+) cells enhances hematopoietic colony formation in vitro, whereas HPIP knockdown leads to a reduction in the number of such colonies. Interestingly, erythroid colony number was significantly higher in HPIP-overexpressing cells. In addition, forced expression of HPIP in K562 cells, a multipotent erythro-megakaryoblastic leukemia cell line, led to an induction of erythroid differentiation. HPIP overexpression in both CD34(+) and K562 cells was associated with increased activation of the PI3K/AKT pathway, and corresponding treatment with a PI3K-specific inhibitor, LY-294002, caused a reduction in clonogenic progenitor number in HPIP-expressing CD34(+) cells and decreased K562 cell differentiation. Combined, these findings point to an important role of the PI3K/AKT pathway in mediating HPIP-induced effects on the growth and differentiation of hematopoietic cells. Interestingly, HPIP gene expression was found to be induced in K562 cells in response to erythroid differentiation signals such as DMSO and erythropoietin. The erythroid lineage-specific transcription factor GATA1 binds to the HPIP promoter and activates HPIP gene transcription in a CCCTC-binding factor (CTCF)-dependent manner. Co-immunoprecipitation and co-localization experiments revealed the association of CTCF with GATA1 indicating the recruitment of CTCF/GATA1 transcription factor complex onto the HPIP promoter. Together, this study provides evidence that HPIP is a target of GATA1 and CTCF in erythroid cells and plays an important role in erythroid differentiation by modulating the PI3K/AKT pathway.
Figures
References
-
- Weissman I. L., Anderson D. J., Gage F. (2001) Stem and progenitor cells. Origins, phenotypes, lineage commitments, and transdifferentiations. Annu. Rev. Cell Dev. Biol. 17, 387–403 - PubMed
-
- Sauvageau G., Iscove N. N., Humphries R. K. (2004) In vitro and in vivo expansion of hematopoietic stem cells. Oncogene 23, 7223–7232 - PubMed
-
- Zon L. I. (2008) Intrinsic and extrinsic control of hematopoietic stem-cell self-renewal. Nature 453, 306–313 - PubMed
-
- Hope K. J., Jin L., Dick J. E. (2003) Human acute myeloid leukemia stem cells. Arch. Med. Res. 34, 507–514 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

