Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
- PMID: 22187439
- PMCID: PMC3281642
- DOI: 10.1074/jbc.M111.321992
Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
Abstract
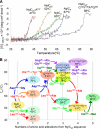
We performed x-ray crystallographic analyses of the 6-aminohexanoate oligomer hydrolase (NylC) from Agromyces sp. at 2.0 Å-resolution. This enzyme is a member of the N-terminal nucleophile hydrolase superfamily that is responsible for the degradation of the nylon-6 industry byproduct. We observed four identical heterodimers (27 kDa + 9 kDa), which resulted from the autoprocessing of the precursor protein (36 kDa) and which constitute the doughnut-shaped quaternary structure. The catalytic residue of NylC was identified as the N-terminal Thr-267 of the 9-kDa subunit. Furthermore, each heterodimer is folded into a single domain, generating a stacked αββα core structure. Amino acid mutations at subunit interfaces of the tetramer were observed to drastically alter the thermostability of the protein. In particular, four mutations (D122G/H130Y/D36A/E263Q) of wild-type NylC from Arthrobacter sp. (plasmid pOAD2-encoding enzyme), with a heat denaturation temperature of T(m) = 52 °C, enhanced the protein thermostability by 36 °C (T(m) = 88 °C), whereas a single mutation (G111S or L137A) decreased the stability by ∼10 °C. We examined the enzymatic hydrolysis of nylon-6 by the thermostable NylC mutant. Argon cluster secondary ion mass spectrometry analyses of the reaction products revealed that the major peak of nylon-6 (m/z 10,000-25,000) shifted to a smaller range, producing a new peak corresponding to m/z 1500-3000 after the enzyme treatment at 60 °C. In addition, smaller fragments in the soluble fraction were successively hydrolyzed to dimers and monomers. Based on these data, we propose that NylC should be designated as nylon hydrolase (or nylonase). Three potential uses of NylC for industrial and environmental applications are also discussed.
Figures

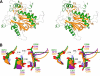



Similar articles
-
X-ray crystallographic analysis of the 6-aminohexanoate cyclic dimer hydrolase: catalytic mechanism and evolution of an enzyme responsible for nylon-6 byproduct degradation.J Biol Chem. 2010 Jan 8;285(2):1239-48. doi: 10.1074/jbc.M109.041285. Epub 2009 Nov 3. J Biol Chem. 2010. PMID: 19889645 Free PMC article.
-
Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase.Sci Rep. 2018 Jun 27;8(1):9725. doi: 10.1038/s41598-018-27860-w. Sci Rep. 2018. PMID: 29950566 Free PMC article.
-
X-ray crystallographic and mutational analysis of the NylC precursor: catalytic mechanism of autocleavage and substrate hydrolysis of nylon hydrolase.FEBS J. 2023 Jul;290(13):3400-3421. doi: 10.1111/febs.16755. Epub 2023 Mar 1. FEBS J. 2023. PMID: 36799721
-
X-ray crystallographic analysis of 6-aminohexanoate-dimer hydrolase: molecular basis for the birth of a nylon oligomer-degrading enzyme.J Biol Chem. 2005 Nov 25;280(47):39644-52. doi: 10.1074/jbc.M505946200. Epub 2005 Sep 14. J Biol Chem. 2005. PMID: 16162506
-
Biodegradation of nylon oligomers.Appl Microbiol Biotechnol. 2000 Oct;54(4):461-6. doi: 10.1007/s002530000434. Appl Microbiol Biotechnol. 2000. PMID: 11092619 Review.
Cited by
-
Metagenomic Exploration of Plastic Degrading Microbes for Biotechnological Application.Curr Genomics. 2020 May;21(4):253-270. doi: 10.2174/1389202921999200525155711. Curr Genomics. 2020. PMID: 33071619 Free PMC article. Review.
-
Natural diversity screening, assay development, and characterization of nylon-6 enzymatic depolymerization.Nat Commun. 2024 Feb 9;15(1):1217. doi: 10.1038/s41467-024-45523-5. Nat Commun. 2024. PMID: 38336849 Free PMC article.
-
Quantitative nylon monomerization by the combination of chemical pretreatment and enzymatic hydrolysis using nylon hydrolases.PLoS One. 2025 Feb 10;20(2):e0318641. doi: 10.1371/journal.pone.0318641. eCollection 2025. PLoS One. 2025. PMID: 39928594 Free PMC article.
-
Targeted Genome Mining Facilitates the Discovery of a Promiscuous, Hyperthermostable Amidase from Thermovenabulum Gondwanense with Notable Nylon-Degrading Capacity.Angew Chem Int Ed Engl. 2025 Jan 10;64(2):e202414842. doi: 10.1002/anie.202414842. Epub 2024 Nov 16. Angew Chem Int Ed Engl. 2025. PMID: 39441552 Free PMC article.
-
Mitigation measures to avert the impacts of plastics and microplastics in the marine environment (a review).Environ Sci Pollut Res Int. 2018 Apr;25(10):9293-9310. doi: 10.1007/s11356-018-1499-z. Epub 2018 Feb 22. Environ Sci Pollut Res Int. 2018. PMID: 29470754
References
-
- Dasgupta S., Hammond W. B., Goddard W. A., 3rd (1996) Crystal structures and properties of nylon polymers from theory. J. Am. Chem. Soc. 118, 12291–12301
-
- Negoro S. (2000) Biodegradation of nylon oligomers. Appl. Microbiol. Biotechnol. 54, 461–466 - PubMed
-
- Okada H., Negoro S., Kimura H., Nakamura S. (1983) Evolutionary adaptation of plasmid-encoded enzymes for degrading nylon oligomers. Nature 306, 203–206 - PubMed
-
- Kato K., Ohtsuki K., Koda Y., Maekawa T., Yomo T., Negoro S., Urabe I. (1995) A plasmid encoding enzymes for nylon oligomer degradation: nucleotide sequence and analysis of pOAD2. Microbiology 141, 2585–2590 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

