Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
- PMID: 22187439
- PMCID: PMC3281642
- DOI: 10.1074/jbc.M111.321992
Three-dimensional structure of nylon hydrolase and mechanism of nylon-6 hydrolysis
Abstract
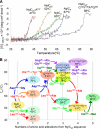
We performed x-ray crystallographic analyses of the 6-aminohexanoate oligomer hydrolase (NylC) from Agromyces sp. at 2.0 Å-resolution. This enzyme is a member of the N-terminal nucleophile hydrolase superfamily that is responsible for the degradation of the nylon-6 industry byproduct. We observed four identical heterodimers (27 kDa + 9 kDa), which resulted from the autoprocessing of the precursor protein (36 kDa) and which constitute the doughnut-shaped quaternary structure. The catalytic residue of NylC was identified as the N-terminal Thr-267 of the 9-kDa subunit. Furthermore, each heterodimer is folded into a single domain, generating a stacked αββα core structure. Amino acid mutations at subunit interfaces of the tetramer were observed to drastically alter the thermostability of the protein. In particular, four mutations (D122G/H130Y/D36A/E263Q) of wild-type NylC from Arthrobacter sp. (plasmid pOAD2-encoding enzyme), with a heat denaturation temperature of T(m) = 52 °C, enhanced the protein thermostability by 36 °C (T(m) = 88 °C), whereas a single mutation (G111S or L137A) decreased the stability by ∼10 °C. We examined the enzymatic hydrolysis of nylon-6 by the thermostable NylC mutant. Argon cluster secondary ion mass spectrometry analyses of the reaction products revealed that the major peak of nylon-6 (m/z 10,000-25,000) shifted to a smaller range, producing a new peak corresponding to m/z 1500-3000 after the enzyme treatment at 60 °C. In addition, smaller fragments in the soluble fraction were successively hydrolyzed to dimers and monomers. Based on these data, we propose that NylC should be designated as nylon hydrolase (or nylonase). Three potential uses of NylC for industrial and environmental applications are also discussed.
Figures

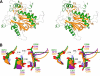



References
-
- Dasgupta S., Hammond W. B., Goddard W. A., 3rd (1996) Crystal structures and properties of nylon polymers from theory. J. Am. Chem. Soc. 118, 12291–12301
-
- Negoro S. (2000) Biodegradation of nylon oligomers. Appl. Microbiol. Biotechnol. 54, 461–466 - PubMed
-
- Okada H., Negoro S., Kimura H., Nakamura S. (1983) Evolutionary adaptation of plasmid-encoded enzymes for degrading nylon oligomers. Nature 306, 203–206 - PubMed
-
- Kato K., Ohtsuki K., Koda Y., Maekawa T., Yomo T., Negoro S., Urabe I. (1995) A plasmid encoding enzymes for nylon oligomer degradation: nucleotide sequence and analysis of pOAD2. Microbiology 141, 2585–2590 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

