Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome
- PMID: 22187675
- PMCID: PMC3264910
- DOI: 10.1261/rna.031187.111
Crystal structure of release factor RF3 trapped in the GTP state on a rotated conformation of the ribosome
Abstract
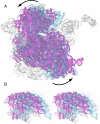
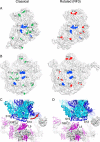
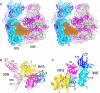
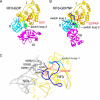
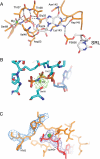
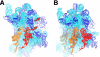
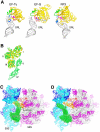
The class II release factor RF3 is a GTPase related to elongation factor EF-G, which catalyzes release of class I release factors RF1 and RF2 from the ribosome after termination of protein synthesis. The 3.3 Å crystal structure of the RF3·GDPNP·ribosome complex provides a high-resolution description of interactions and structural rearrangements that occur when binding of this translational GTPase induces large-scale rotational movements in the ribosome. RF3 induces a 7° rotation of the body and 14° rotation of the head of the 30S ribosomal subunit, and itself undergoes inter- and intradomain conformational rearrangements. We suggest that ordering of critical elements of switch loop I and the P loop, which help to form the GTPase catalytic site, are caused by interactions between the G domain of RF3 and the sarcin-ricin loop of 23S rRNA. The rotational movements in the ribosome induced by RF3, and its distinctly different binding orientation to the sarcin-ricin loop of 23S rRNA, raise interesting implications for the mechanism of action of EF-G in translocation.
Figures

Comment in
-
Another burst of smoke: atomic resolution structures of RF3 bound to the ribosome.RNA. 2012 Apr;18(4):605-9. doi: 10.1261/rna.032011.111. Epub 2012 Feb 17. RNA. 2012. PMID: 22345149 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
