Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling
- PMID: 22187934
- PMCID: PMC3258656
- DOI: 10.1042/BJ20111451
Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling
Abstract
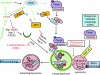
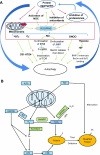
Reactive oxygen and nitrogen species change cellular responses through diverse mechanisms that are now being defined. At low levels, they are signalling molecules, and at high levels, they damage organelles, particularly the mitochondria. Oxidative damage and the associated mitochondrial dysfunction may result in energy depletion, accumulation of cytotoxic mediators and cell death. Understanding the interface between stress adaptation and cell death then is important for understanding redox biology and disease pathogenesis. Recent studies have found that one major sensor of redox signalling at this switch in cellular responses is autophagy. Autophagic activities are mediated by a complex molecular machinery including more than 30 Atg (AuTophaGy-related) proteins and 50 lysosomal hydrolases. Autophagosomes form membrane structures, sequester damaged, oxidized or dysfunctional intracellular components and organelles, and direct them to the lysosomes for degradation. This autophagic process is the sole known mechanism for mitochondrial turnover. It has been speculated that dysfunction of autophagy may result in abnormal mitochondrial function and oxidative or nitrative stress. Emerging investigations have provided new understanding of how autophagy of mitochondria (also known as mitophagy) is controlled, and the impact of autophagic dysfunction on cellular oxidative stress. The present review highlights recent studies on redox signalling in the regulation of autophagy, in the context of the basic mechanisms of mitophagy. Furthermore, we discuss the impact of autophagy on mitochondrial function and accumulation of reactive species. This is particularly relevant to degenerative diseases in which oxidative stress occurs over time, and dysfunction in both the mitochondrial and autophagic pathways play a role.
Figures

References
-
- de Duve C. The lysosome. Sci. Am. 1963;208:64–72. - PubMed
-
- de Duve C., Wattiaux R. Functions of lysosomes. Annu. Rev. Physiol. 1966;28:435–492. - PubMed
-
- Shacka J. J., Roth K. A., Zhang J. The autophagy–lysosomal degradation pathway: role in neurodegenerative disease and therapy. Front. Biosci. 2008;13:718–736. - PubMed
-
- Crifo C., Capuozzo E., Siems W., Salerno C. Inhibition of ion transport ATPases by HNE. Biofactors. 2005;24:137–140. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

