Cdx2 homeoprotein inhibits non-homologous end joining in colon cancer but not in leukemia cells
- PMID: 22189105
- PMCID: PMC3333856
- DOI: 10.1093/nar/gkr1242
Cdx2 homeoprotein inhibits non-homologous end joining in colon cancer but not in leukemia cells
Abstract
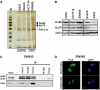
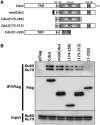
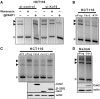
Cdx2, a gene of the paraHox cluster, encodes a homeodomain transcription factor that plays numerous roles in embryonic development and in homeostasis of the adult intestine. Whereas Cdx2 exerts a tumor suppressor function in the gut, its abnormal ectopic expression in acute leukemia is associated to a pro-oncogenic function. To try to understand this duality, we have hypothesized that Cdx2 may interact with different protein partners in the two tissues and set up experiments to identify them by tandem affinity purification. We show here that Cdx2 interacts with the Ku heterodimer specifically in intestinal cells, but not in leukemia cells, via its homeodomain. Ku proteins do not affect Cdx2 transcriptional activity. However, Cdx2 inhibits in vivo and in vitro the DNA repair activity mediated by Ku proteins in intestinal cells. Whereas Cdx2 does not affect the recruitment of Ku proteins and DNA-PKcs into the DNA repair complex, it inhibits DNA-PKcs activity. Thus, we report here a new function of Cdx2, acting as an inhibitor of the DNA repair machinery, that may contribute to its tumor suppressor function specifically in the gut.
Figures
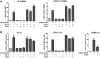

Similar articles
-
Extending the functions of the homeotic transcription factor Cdx2 in the digestive system through nontranscriptional activities.World J Gastroenterol. 2015 Feb 7;21(5):1436-43. doi: 10.3748/wjg.v21.i5.1436. World J Gastroenterol. 2015. PMID: 25663763 Free PMC article. Review.
-
Distinct mechanisms for opposite functions of homeoproteins Cdx2 and HoxB7 in double-strand break DNA repair in colon cancer cells.Cancer Lett. 2016 May 1;374(2):208-15. doi: 10.1016/j.canlet.2016.02.026. Epub 2016 Feb 19. Cancer Lett. 2016. PMID: 26902420
-
An Intrinsically Disordered APLF Links Ku, DNA-PKcs, and XRCC4-DNA Ligase IV in an Extended Flexible Non-homologous End Joining Complex.J Biol Chem. 2016 Dec 30;291(53):26987-27006. doi: 10.1074/jbc.M116.751867. Epub 2016 Nov 14. J Biol Chem. 2016. PMID: 27875301 Free PMC article.
-
The binding of Ku antigen to homeodomain proteins promotes their phosphorylation by DNA-dependent protein kinase.J Biol Chem. 2001 May 18;276(20):16848-56. doi: 10.1074/jbc.M100768200. Epub 2001 Feb 20. J Biol Chem. 2001. PMID: 11279128
-
The Ku heterodimer: function in DNA repair and beyond.Mutat Res Rev Mutat Res. 2015 Jan-Mar;763:15-29. doi: 10.1016/j.mrrev.2014.06.002. Epub 2014 Jul 4. Mutat Res Rev Mutat Res. 2015. PMID: 25795113 Review.
Cited by
-
Homeobox Gene Deregulation: Impact on the Hallmarks of Cancer.Cancer Hallm. 2013 Sep 1;1(2-3):67-76. doi: 10.1166/ch.2013.1007. Cancer Hallm. 2013. PMID: 24761365 Free PMC article.
-
Dual functions of the homeoprotein DLX4 in modulating responsiveness of tumor cells to topoisomerase II-targeting drugs.Cancer Res. 2013 Jan 15;73(2):1000-10. doi: 10.1158/0008-5472.CAN-12-3538. Epub 2012 Dec 7. Cancer Res. 2013. PMID: 23222298 Free PMC article.
-
Extending the functions of the homeotic transcription factor Cdx2 in the digestive system through nontranscriptional activities.World J Gastroenterol. 2015 Feb 7;21(5):1436-43. doi: 10.3748/wjg.v21.i5.1436. World J Gastroenterol. 2015. PMID: 25663763 Free PMC article. Review.
-
CDX1 and CDX2 suppress colon cancer stemness by inhibiting β-catenin-facilitated formation of Pol II-DSIF-PAF1C complex.Cell Death Dis. 2025 May 21;16(1):408. doi: 10.1038/s41419-025-07737-3. Cell Death Dis. 2025. PMID: 40399276 Free PMC article.
-
Homeodomain Proteins Directly Regulate ATM Kinase Activity.Cell Rep. 2018 Aug 7;24(6):1471-1483. doi: 10.1016/j.celrep.2018.06.089. Cell Rep. 2018. PMID: 30089259 Free PMC article.
References
-
- Chawengsaksophak K, James R, Hammond VE, Kontgen F, Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–87. - PubMed
-
- Van Den Akker E, Forlani S, Chawengsaksophak K, De Graaff W, Beck F, Meyer BI, Deschamps J. Cdx1 and Cdx2 have overlapping functions in anteroposterior patterning and posterior axis elongation. Development. 2002;129:2181–2193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

