The BRAF(V600E) causes widespread alterations in gene methylation in the genome of melanoma cells
- PMID: 22189819
- PMCID: PMC3293379
- DOI: 10.4161/cc.11.2.18707
The BRAF(V600E) causes widespread alterations in gene methylation in the genome of melanoma cells
Abstract
Although BRAF(V600E) is well known to play an important role in the tumorigenesis of melanoma, its molecular mechanism, particularly the epigenetic aspect, has been incompletely understood. Here, we investigated the role of BRAF(V600E) signaling in altering gene methylation in the genome of melanoma cells using a methylated CpG island amplification/CpG island microarray system and searched for genes coupled to the BRAF(V600E) signaling through methylation aberrations. The results indicated that a wide range of genes with broad functions were linked to BRAF(V600E) signaling through their hyper- or hypomethylation. Expression of 59 genes hypermethylated upon BRAF knockdown was selectively tested and found to be largely correspondingly underexpressed, suggesting that these genes were naturally hypomethylated, and overexpressed with BRAF(V600E) in melanoma. This BRAF(V600E)-promoted hypomethylation was confirmed on genes selectively examined in primary melanoma tumors. Some of these genes were functionally tested and demonstrated to play a role in melanoma cell proliferation and invasion. As a mechanism of aberrant gene methylation driven by BRAF(V600E), expression of the DNA methyltransferase 1 and histone methyltransferase EZH2 was profoundly affected by BRAF(V600E). We have thus uncovered a previously unrecognized prominent epigenetic mechanism in the tumorigenesis of melanoma driven by BRAF(V600E). Many of the functionally important genes controlled by the BRAF(V600E) signaling through aberrant methylation may prove to be novel therapeutic targets for melanoma.
Figures

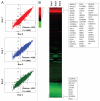


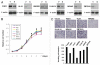
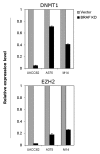
Comment in
-
BRAF mutation: a double-edged sword in epigenetic alterations?Cell Cycle. 2012 Feb 1;11(3):434-5. doi: 10.4161/cc.11.3.19170. Epub 2012 Feb 1. Cell Cycle. 2012. PMID: 22262188 No abstract available.
Similar articles
-
Genome-wide alterations in gene methylation by the BRAF V600E mutation in papillary thyroid cancer cells.Endocr Relat Cancer. 2011 Nov 14;18(6):687-97. doi: 10.1530/ERC-11-0212. Print 2011 Dec. Endocr Relat Cancer. 2011. PMID: 21937738 Free PMC article.
-
In-depth genomic data analyses revealed complex transcriptional and epigenetic dysregulations of BRAFV600E in melanoma.Mol Cancer. 2015 Mar 14;14:60. doi: 10.1186/s12943-015-0328-y. Mol Cancer. 2015. PMID: 25890285 Free PMC article.
-
Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling.PLoS One. 2009 Dec 21;4(12):e8357. doi: 10.1371/journal.pone.0008357. PLoS One. 2009. PMID: 20027224 Free PMC article.
-
Epigenetic regulation in human melanoma: past and future.Epigenetics. 2015;10(2):103-21. doi: 10.1080/15592294.2014.1003746. Epigenetics. 2015. PMID: 25587943 Free PMC article. Review.
-
Multifaceted Roles of DNA Methylation in Neoplastic Transformation, from Tumor Suppressors to EMT and Metastasis.Genes (Basel). 2020 Aug 12;11(8):922. doi: 10.3390/genes11080922. Genes (Basel). 2020. PMID: 32806509 Free PMC article. Review.
Cited by
-
Epigenetic genes regulated by the BRAFV600E signaling are associated with alterations in the methylation and expression of tumor suppressor genes and patient survival in melanoma.Biochem Biophys Res Commun. 2012 Aug 17;425(1):45-50. doi: 10.1016/j.bbrc.2012.07.046. Epub 2012 Jul 17. Biochem Biophys Res Commun. 2012. PMID: 22820187 Free PMC article.
-
Extracellular Vesicles and Epigenetic Modifications Are Hallmarks of Melanoma Progression.Int J Mol Sci. 2019 Dec 20;21(1):52. doi: 10.3390/ijms21010052. Int J Mol Sci. 2019. PMID: 31861757 Free PMC article. Review.
-
DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: mechanism and clinical application.Clin Epigenetics. 2021 Aug 27;13(1):166. doi: 10.1186/s13148-021-01154-x. Clin Epigenetics. 2021. PMID: 34452630 Free PMC article. Review.
-
EZH2 as a mediator of treatment resistance in melanoma.Pigment Cell Melanoma Res. 2016 Sep;29(5):500-7. doi: 10.1111/pcmr.12481. Epub 2016 May 25. Pigment Cell Melanoma Res. 2016. PMID: 27063195 Free PMC article. Review.
-
Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy.Cancer Discov. 2014 Jan;4(1):80-93. doi: 10.1158/2159-8290.CD-13-0642. Epub 2013 Nov 21. Cancer Discov. 2014. PMID: 24265155 Free PMC article.
References
-
- Altekruse SF, Kosary CL, Krapcho M, Neyman N, Aminou R, Waldron W, et al. SEER Cancer Statistics Review, 1975–2007. Bethesda, MD: National Cancer Institute; http://seer.cancer.gov/csr/1975_2007/, based on November 2009 SEER data submission, posted to the SEER web site 2010.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
