Molecular epidemiology of HIV in two highly endemic areas of northeastern South Africa
- PMID: 22189822
- PMCID: PMC4957143
- DOI: 10.1007/s00705-011-1180-z
Molecular epidemiology of HIV in two highly endemic areas of northeastern South Africa
Abstract
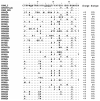
There is paucity of data on the genetic landscape of HIV-1 viruses circulating in the Limpopo Province of northeastern South Africa. Here, we examine the genetic diversity of viruses from Bela-Bela and Musina, two towns with high HIV prevalence. Between June 2007 and March 2008, blood samples were collected from antiretroviral-drug-naïve individuals. Viruses were analyzed for genetic subtypes and drug resistance mutations. All of the viruses in these samples were shown by phylogenetic analysis based on gag p17, gag p24, reverse transcriptase, protease and envelope C2-C3 gene regions to belong to HIV-1 subtype C. Two of 44 reverse transcriptase sequences (4.5%) contained N rather than the consensus K at position 103. The K103N mutation is normally associated with resistance to NNRTIs. No major mutations were observed in the protease gene. However, several polymorphisms and amino acid changes normally considered to be minor drug resistance mutations were observed in the protease sequences. These results suggest that HIV-1 subtype C remains the predominant variant responsible for the epidemic in northeastern South Africa and that the prevalence of drug-resistant viruses among the naïve population is low.
Figures



References
-
- Simon-Loriere E, Rossolillo P, Negroni M. RNA structures, genomic organization and selection of recombinant HIV. RNA Biol. 2011;8(2):280–286. - PubMed
-
- Powell RL, Zhao J, Konings FA, et al. Circulating recombinant form (CRF) 37_cpx: an old strain in Cameroon composed of diverse, genetically distant lineages of subtypes A and G. AIDS Res Hum Retroviruses. 2007;23(7):923–933. - PubMed
-
- Brennan CA, Brites C, Bodelle P, et al. HIV-1 strains identified in Brazilian blood donors: significant prevalence of B/F1 recombinants. AIDS Res Hum Retroviruses. 2007;23(11):1434–1441. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

